Dependence of paracentric inversion rate on tract length
- PMID: 17407601
- PMCID: PMC1858705
- DOI: 10.1186/1471-2105-8-115
Dependence of paracentric inversion rate on tract length
Abstract
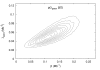
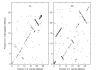
Background: We develop a Bayesian method based on MCMC for estimating the relative rates of pericentric and paracentric inversions from marker data from two species. The method also allows estimation of the distribution of inversion tract lengths.
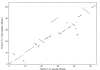
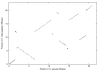
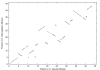
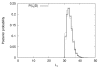
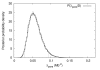
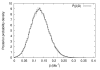
Results: We apply the method to data from Drosophila melanogaster and D. yakuba. We find that pericentric inversions occur at a much lower rate compared to paracentric inversions. The average paracentric inversion tract length is approx. 4.8 Mb with small inversions being more frequent than large inversions. If the two breakpoints defining a paracentric inversion tract are uniformly and independently distributed over chromosome arms there will be more short tract-length inversions than long; we find an even greater preponderance of short tract lengths than this would predict. Thus there appears to be a correlation between the positions of breakpoints which favors shorter tract lengths.
Conclusion: The method developed in this paper provides the first statistical estimator for estimating the distribution of inversion tract lengths from marker data. Application of this method for a number of data sets may help elucidate the relationship between the length of an inversion and the chance that it will get accepted.
Figures
References
-
- Watterson GA, Ewens WJ, Hall TE, Morgan A. The chromosome inversion problem. Journal of Theoretical Biology. 1982;99:1–7. doi: 10.1016/0022-5193(82)90384-8. - DOI
-
- Kececioglu J, Sankoff D. Exact and approximation algorithms for the inversion distance between two permutations. Algortihmica. 1995;13:180–210. doi: 10.1007/BF01188586. - DOI
-
- Hannenhalli S, Pevzner PA. Transforming cabbage into turnip (polynomial algorithm for sorting signed permutations by reversals) Proceedings of the 27th Annual ACM Symposium on the Theory of Computing. 1995. pp. 1–27.
-
- Larget B, Simon DL, Kadane JB. Bayesian phylogenetic inference from animal mitochondrial genome arrangements (with discussion) J R Stat Soc Ser B. 2002;64:681–693. doi: 10.1111/1467-9868.00356. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

