Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse
- PMID: 17407603
- PMCID: PMC1855325
- DOI: 10.1186/1471-2164-8-92
Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse
Abstract
Background: Neoplasia can be driven by mutations resulting in dysregulation of transcription. In the mesenchymal neoplasm, aggressive fibromatosis, subtractive hybridization identified sterile alpha motif domain 9 (SAMD9) as a substantially down regulated gene in neoplasia. SAMD9 was recently found to be mutated in normophosphatemic familial tumoral calcinosis. In this study, we studied the gene structure and function of SAMD9, and its paralogous gene, SAMD9L, and examined these in a variety of species.
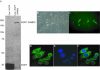
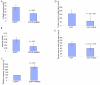
Results: SAMD9 is located on human chromosome 7q21.2 with a paralogous gene sterile alpha motif domain 9 like (SAMD9L) in the head-to-tail orientation. Although both genes are present in a variety of species, the orthologue for SAMD9 is lost in the mouse lineage due to a unique genomic rearrangement. Both SAMD9 and SAMD9L are ubiquitously expressed in human tissues. SAMD9 is expressed at a lower level in a variety of neoplasms associated with beta-catenin stabilization, such as aggressive fibromatosis, breast, and colon cancers. SAMD9 and SAMD9L contain an amino-terminal SAM domain, but the remainder of the predicted protein structure does not exhibit substantial homology to other known protein motifs. The putative protein product of SAMD9 localizes to the cytoplasm. In vitro data shows that SAMD9 negatively regulates cell proliferation. Over expression of SAMD9 in the colon cancer cell line, SW480, reduces the volume of tumors formed when transplanted into immune-deficient mice.
Conclusion: SAMD9 and SAMD9L are a novel family of genes, which play a role regulating cell proliferation and suppressing the neoplastic phenotype. This is the first report as far as we know about a human gene that exists in rat, but is lost in mouse, due to a mouse specific rearrangement, resulting in the loss of the SAMD9 gene.
Figures





References
-
- Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kuhl M, Behrens J, von der Mark K, Starzinski-Powitz A, Wixler V. The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J Cell Biol. 2002;159:113–122. doi: 10.1083/jcb.200202075. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

