Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression
- PMID: 17408486
- PMCID: PMC1896000
- DOI: 10.1186/gb-2007-8-4-r49
Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression
Abstract
Background: Arabidopsis thaliana transcript profiles indicate effects of abiotic and biotic stresses and tissue-specific and cell-specific gene expression. Organizing these datasets could reveal the structure and mechanisms of responses and crosstalk between pathways, and in which cells the plants perceive, signal, respond to, and integrate environmental inputs.
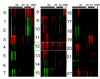
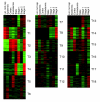
Results: We clustered Arabidopsis transcript profiles for various treatments, including abiotic, biotic, and chemical stresses. Ubiquitous stress responses in Arabidopsis, similar to those of fungi and animals, employ genes in pathways related to mitogen-activated protein kinases, Snf1-related kinases, vesicle transport, mitochondrial functions, and the transcription machinery. Induced responses to stresses are attributed to genes whose promoters are characterized by a small number of regulatory motifs, although secondary motifs were also apparent. Most genes that are downregulated by stresses exhibited distinct tissue-specific expression patterns and appear to be under developmental regulation. The abscisic acid-dependent transcriptome is delineated in the cluster structure, whereas functions that are dependent on reactive oxygen species are widely distributed, indicating that evolutionary pressures confer distinct responses to different stresses in time and space. Cell lineages in roots express stress-responsive genes at different levels. Intersections of stress-responsive and cell-specific profiles identified cell lineages affected by abiotic stress.
Conclusion: By analyzing the stress-dependent expression profile, we define a common stress transcriptome that apparently represents universal cell-level stress responses. Combining stress-dependent and tissue-specific and cell-specific expression profiles, and Arabidopsis 5'-regulatory DNA sequences, we confirm known stress-related 5' cis-elements on a genome-wide scale, identify secondary motifs, and place the stress response within the context of tissues and cell lineages in the Arabidopsis root.
Figures





References
-
- Chinnusamy V, Stevenson B, Lee BH, Zhu JK. Screening for gene regulation mutants by bioluminescence imaging. Sci STKE. 2002;2002:PL10. - PubMed
-
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

