Genetic microheterogeneity and phenotypic variation of Helicobacter pylori arginase in clinical isolates
- PMID: 17408487
- PMCID: PMC1853099
- DOI: 10.1186/1471-2180-7-26
Genetic microheterogeneity and phenotypic variation of Helicobacter pylori arginase in clinical isolates
Abstract
Background: Clinical isolates of the gastric pathogen Helicobacter pylori display a high level of genetic macro- and microheterogeneity, featuring a panmictic, rather than clonal structure. The ability of H. pylori to survive the stomach acid is due, in part, to the arginase-urease enzyme system. Arginase (RocF) hydrolyzes L-arginine to L-ornithine and urea, and urease hydrolyzes urea to carbon dioxide and ammonium, which can neutralize acid.
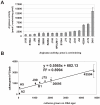
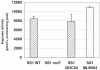
Results: The degree of variation in arginase was explored at the DNA sequence, enzyme activity and protein expression levels. To this end, arginase activity was measured from 73 minimally-passaged clinical isolates and six laboratory-adapted strains of H. pylori. The rocF gene from 21 of the strains was cloned into genetically stable E. coli and the enzyme activities measured. Arginase activity was found to substantially vary (>100-fold) in both different H. pylori strains and in the E. coli model. Western blot analysis revealed a positive correlation between activity and amount of protein expressed in most H. pylori strains. Several H. pylori strains featured altered arginase activity upon in vitro passage. Pairwise alignments of the 21 rocF genes plus strain J99 revealed extensive microheterogeneity in the promoter region and 3' end of the rocF coding region. Amino acid S232, which was I232 in the arginase-negative clinical strain A2, was critical for arginase activity.
Conclusion: These studies demonstrated that H. pylori arginase exhibits extensive genotypic and phenotypic variation which may be used to understand mechanisms of microheterogeneity in H. pylori.
Figures




References
-
- Blaser MJ. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology. 1987;93:371–383. - PubMed
-
- Blaser MJ. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. Journal of Infectious Diseases. 1990;161:626–633. - PubMed
-
- Marshall BJ, McGechie DB, Rogers PA, Glancy RJ. Pyloric Campylobacter infection and gastroduodenal disease. Medical Journal of Australia. 1985;142:439–444. - PubMed
-
- Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii.[see comment] New England Journal of Medicine. 1991;325:1132–1136. - PubMed
-
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

