Accurate, precise modeling of cell proliferation kinetics from time-lapse imaging and automated image analysis of agar yeast culture arrays
- PMID: 17408510
- PMCID: PMC1847469
- DOI: 10.1186/1752-0509-1-3
Accurate, precise modeling of cell proliferation kinetics from time-lapse imaging and automated image analysis of agar yeast culture arrays
Abstract
Background: Genome-wide mutant strain collections have increased demand for high throughput cellular phenotyping (HTCP). For example, investigators use HTCP to investigate interactions between gene deletion mutations and additional chemical or genetic perturbations by assessing differences in cell proliferation among the collection of 5000 S. cerevisiae gene deletion strains. Such studies have thus far been predominantly qualitative, using agar cell arrays to subjectively score growth differences. Quantitative systems level analysis of gene interactions would be enabled by more precise HTCP methods, such as kinetic analysis of cell proliferation in liquid culture by optical density. However, requirements for processing liquid cultures make them relatively cumbersome and low throughput compared to agar. To improve HTCP performance and advance capabilities for quantifying interactions, YeastXtract software was developed for automated analysis of cell array images.
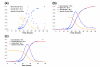
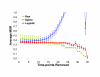
Results: YeastXtract software was developed for kinetic growth curve analysis of spotted agar cultures. The accuracy and precision for image analysis of agar culture arrays was comparable to OD measurements of liquid cultures. Using YeastXtract, image intensity vs. biomass of spot cultures was linearly correlated over two orders of magnitude. Thus cell proliferation could be measured over about seven generations, including four to five generations of relatively constant exponential phase growth. Spot area normalization reduced the variation in measurements of total growth efficiency. A growth model, based on the logistic function, increased precision and accuracy of maximum specific rate measurements, compared to empirical methods. The logistic function model was also more robust against data sparseness, meaning that less data was required to obtain accurate, precise, quantitative growth phenotypes.
Conclusion: Microbial cultures spotted onto agar media are widely used for genotype-phenotype analysis, however quantitative HTCP methods capable of measuring kinetic growth rates have not been available previously. YeastXtract provides objective, automated, quantitative, image analysis of agar cell culture arrays. Fitting the resulting data to a logistic equation-based growth model yields robust, accurate growth rate information. These methods allow the incorporation of imaging and automated image analysis of cell arrays, grown on solid agar media, into HTCP-driven experimental approaches, such as global, quantitative analysis of gene interaction networks.
Figures






References
-
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2003 - PubMed
-
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

