Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae
- PMID: 17409072
- PMCID: PMC1894612
- DOI: 10.1534/genetics.106.069732
Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae
Abstract
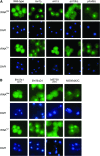
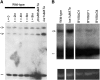
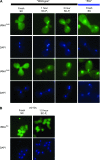
Nuclear export of tRNA is an essential eukaryotic function, yet the one known yeast tRNA nuclear exporter, Los1, is nonessential. Moreover recent studies have shown that tRNAs can move retrograde from the cytosol to the nucleus by an undefined process. Therefore, additional gene products involved in tRNA nucleus-cytosol dynamics have yet to be identified. Synthetic genetic array (SGA) analysis was employed to identify proteins involved in Los1-independent tRNA transport and in regulating tRNA nucleus-cytosol distribution. These studies uncovered synthetic interactions between los1Delta and pho88Delta involved in inorganic phopsphate uptake. Further analysis revealed that inorganic phosphate deprivation causes transient, temperature-dependent nuclear accumulation of mature cytoplasmic tRNA within nuclei via a Mtr10- and retrograde-dependent pathway, providing a novel connection between tRNA subcellular dynamics and phosphate availability.
Figures


References
-
- Arts, G. J., M. Fornerod and I. W. Mattaj, 1998. a Identification of a nuclear export receptor for tRNA. Curr. Biol. 8 305–314. - PubMed
-
- Byrne, M., N. Miller, M. Springer and E. K. O'Shea, 2004. A distal, high-affinity binding site on the cyclin-CDK substrate Pho4 is important for its phosphorylation and regulation. J. Mol. Biol. 335 57–70. - PubMed
-
- Carroll, A. S., and E. K. O'Shea, 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27 87–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

