Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations
- PMID: 17409084
- PMCID: PMC1894625
- DOI: 10.1534/genetics.106.067223
Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations
Abstract
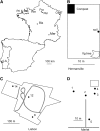
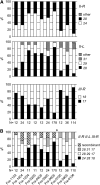
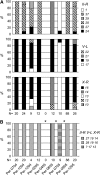
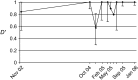
Caenorhabditis elegans is a major laboratory model system yet a newcomer to the field of population genetics, and relatively little is known of its biology in the wild. Recent studies of natural populations at a single time point revealed strong spatial population structure and suggested that these populations may be very dynamic. We have therefore studied several natural C. elegans populations over time and genotyped them at polymorphic microsatellite loci. While some populations appear to be genetically stable over the course of observation, others seem to go extinct, with full replacement of multilocus genotypes upon regrowth. The frequency of heterozygotes indicates that outcrossing occurs at a mean frequency of 1.7% and is variable between populations. However, in genetically stable populations, linkage disequilibrium between different chromosomes can be maintained over several years at a level much higher than expected from the heterozygote frequency. C. elegans seems to follow metapopulation dynamics, and the maintenance of linkage disequilibrium despite a low yet significant level of outcrossing suggests that selection may act against the progeny of outcrossings.
Figures

References
-
- Agrawal, A. F., 2006. Evolution of sex: Why do organisms shuffle their genotypes? Curr. Biol. 16 R696–R704. - PubMed
-
- Barrière, A., and M.-A. Félix, 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15 1176–1184. - PubMed
-
- Barrière, A., and M.-A. Félix, 2006. Isolation of C. elegans and related nematodes in WormBook, edited by The C. elegans Research Community (http://www.wormbook.org/). - PMC - PubMed
-
- Charbonnel, N., and J. Pemberton, 2005. A long-term genetic survey of an ungulate population reveals balancing selection acting on MHC through spatial and temporal fluctuations in selection. Heredity 95 377–388. - PubMed

