Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus
- PMID: 17409141
- PMCID: PMC1900078
- DOI: 10.1128/JVI.00210-07
Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus
Abstract
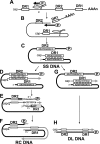
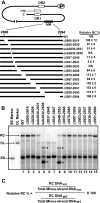
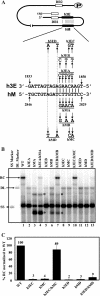
Hepadnaviruses utilize two template switches (primer translocation and circularization) during synthesis of plus-strand DNA to generate a relaxed-circular (RC) DNA genome. In duck hepatitis B virus (DHBV) three cis-acting sequences, 3E, M, and 5E, contribute to both template switches through base pairing, 3E with the 3' portion of M and 5E with the 5' portion of M. Human hepatitis B virus (HBV) also contains multiple cis-acting sequences that contribute to the accumulation of RC DNA, but the mechanisms through which these sequences contribute were previously unknown. Three of the HBV cis-acting sequences (h3E, hM, and h5E) occupy positions equivalent to those of the DHBV 3E, M, and 5E. We present evidence that h3E and hM contribute to the synthesis of RC DNA through base pairing during both primer translocation and circularization. Mutations that disrupt predicted base pairing inhibit both template switches while mutations that restore the predicted base pairing restore function. Therefore, the h3E-hM base pairing appears to be a conserved requirement for template switching during plus-strand DNA synthesis of HBV and DHBV. Also, we show that base pairing is not sufficient to explain the mechanism of h3E and hM, as mutating sequences adjacent to the base pairing regions inhibited both template switches. Finally, we did not identify predicted base pairing between h5E and the hM region, indicating a possible difference between HBV and DHBV. The significance of these similarities and differences between HBV and DHBV will be discussed.
Figures
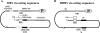

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

