Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase
- PMID: 17409142
- PMCID: PMC1900118
- DOI: 10.1128/JVI.02252-06
Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase
Abstract
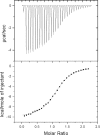
Two critical interactions within the poliovirus RNA replication complex are those of the RNA-dependent RNA polymerase 3D with the viral proteins 3AB and VPg. 3AB is a membrane-binding protein responsible for the localization of the polymerase to the membranous vesicles at which replication occurs. VPg (a peptide comprising the 3B region of 3AB) is the 22-residue soluble product of 3AB cleavage and serves as the protein primer for RNA replication. The detailed interactions of these proteins with the RNA-dependent RNA polymerase 3D were analyzed to elucidate the precise roles of 3AB and VPg in the viral RNA replication complex. Using a membrane-based pull-down assay, we have identified a binding "hot-spot" spanning residues 100 to 104 in the 3B (VPg) region of 3AB which plays a critical role in mediating the interaction of 3AB with the polymerase. Isothermal titration calorimetry shows that the interaction of VPg with 3D is enthalpically driven, with a dissociation constant of 11 microM. Mutational analyses of VPg indicate that a subset of the residues important for 3AB-3D binding are also important for VPg-3D binding. Two residues in particular, P14 and R17, were shown to be absolutely critical for the binding interaction. This work provides the direct characterization of two binding interactions critical for the replication of this important class of viruses and identifies a conserved polymerase binding sequence responsible for targeting the polymerase.
Figures





Similar articles
-
Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer.J Mol Biol. 2006 Mar 24;357(2):665-75. doi: 10.1016/j.jmb.2005.12.044. Epub 2006 Jan 5. J Mol Biol. 2006. PMID: 16427083
-
Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB.J Virol. 2007 Jun;81(11):5669-84. doi: 10.1128/JVI.02350-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360746 Free PMC article.
-
Live cell imaging of protein interactions in poliovirus RNA replication complex using fluorescence resonance energy transfer (FRET).Biochem Biophys Res Commun. 2008 Apr 11;368(3):489-94. doi: 10.1016/j.bbrc.2008.01.094. Epub 2008 Feb 4. Biochem Biophys Res Commun. 2008. PMID: 18252199
-
Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase.Curr Opin Struct Biol. 2009 Dec;19(6):752-8. doi: 10.1016/j.sbi.2009.10.016. Epub 2009 Nov 13. Curr Opin Struct Biol. 2009. PMID: 19914060 Review.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
Cited by
-
Novel roles of the picornaviral 3D polymerase in viral pathogenesis.Adv Virol. 2010 Jan 1;2010:368068. doi: 10.1155/2010/368068. Adv Virol. 2010. PMID: 20625447 Free PMC article.
-
NMR solution structure of poliovirus uridylyated peptide linked to the genome (VPgpU).Peptides. 2010 Aug;31(8):1441-8. doi: 10.1016/j.peptides.2010.04.021. Epub 2010 May 2. Peptides. 2010. PMID: 20441784 Free PMC article.
-
Transgenic expression of the 3D polymerase inhibits Theiler's virus infection and demyelination.J Virol. 2009 Dec;83(23):12279-89. doi: 10.1128/JVI.00664-09. Epub 2009 Sep 16. J Virol. 2009. PMID: 19759133 Free PMC article.
-
Picornaviruses: A View from 3A.Viruses. 2021 Mar 11;13(3):456. doi: 10.3390/v13030456. Viruses. 2021. PMID: 33799649 Free PMC article. Review.
-
Dual role of the foot-and-mouth disease virus 3B1 protein in the replication complex: As protein primer and as an essential component to recruit 3Dpol to membranes.PLoS Pathog. 2023 May 1;19(5):e1011373. doi: 10.1371/journal.ppat.1011373. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37126532 Free PMC article.
References
-
- Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. - PubMed
-
- Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18-29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

