Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression
- PMID: 17409157
- PMCID: PMC1900110
- DOI: 10.1128/JVI.00022-07
Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression
Abstract
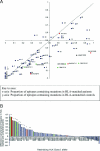
The possession of some HLA class I molecules is associated with delayed progression to AIDS. The mechanism behind this beneficial effect is unclear. We tested the idea that cytotoxic T-cell responses restricted by advantageous HLA class I molecules impose stronger selection pressures than those restricted by other HLA class I alleles. As a measure of the selection pressure imposed by HLA class I alleles, we determined the extent of HLA class I-associated epitope variation in a cohort of European human immunodeficiency virus (HIV)-positive individuals (n=84). We validated our findings in a second, distinct cohort of African patients (n=516). We found that key HIV epitopes restricted by advantageous HLA molecules (B27, B57, and B51 in European patients and B5703, B5801, and B8101 in African patients) were more frequently mutated in individuals bearing the restricting HLA than in those who lacked the restricting HLA class I molecule. HLA alleles associated with clinical benefit restricted certain epitopes for which the consensus peptides were frequently recognized by the immune response despite the circulating virus's being highly polymorphic. We found a significant inverse correlation between the HLA-associated hazard of disease progression and the mean HLA-associated prevalence of mutations within epitopes (P=0.028; R2=0.34). We conclude that beneficial HLA class I alleles impose strong selection at key epitopes. This is revealed by the frequent association between effective T-cell responses and circulating viral escape mutants and the rarity of these variants in patients who lack these favorable HLA class I molecules, suggesting a significant pressure to revert.
Figures




References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLOS Med. 3:e403. doi:10.1371/journal.pmed.0030403. - DOI - PMC - PubMed
-
- Deeks, S. G., J. D. Barbour, J. N. Martin, M. S. Swanson, and R. M. Grant. 2000. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J. Infect. Dis. 181:946-953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

