Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila
- PMID: 17409186
- PMCID: PMC1871816
- DOI: 10.1073/pnas.0701572104
Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila
Abstract
We have estimated the selective effects of amino acid replacements in natural populations by comparing levels of polymorphism in 91 genes in African populations of Drosophila melanogaster with their divergence from Drosophila simulans. The genes include about equal numbers whose level of expression in adults is greater in males, greater in females, or approximately equal in the sexes. Markov chain Monte Carlo methods were used to sample key parameters in the stationary distribution of polymorphism and divergence in a model in which the selective effect of each nonsynonymous mutation is regarded as a random sample from some underlying normal distribution whose mean may differ from one gene to the next. Our analysis suggests that approximately 95% of all nonsynonymous mutations that could contribute to polymorphism or divergence are deleterious, and that the average proportion of deleterious amino acid polymorphisms in samples is approximately 70%. On the other hand, approximately 95% of fixed differences between species are positively selected, although the scaled selection coefficient (N(e)s) is very small. We estimate that approximately 46% of amino acid replacements have N(e)s < 2, approximately 84% have N(e)s < 4, and approximately 99% have N(e)s < 7. Although positive selection among amino acid differences between species seems pervasive, most of the selective effects could be regarded as nearly neutral. There are significant differences in selection between sex-biased and unbiased genes, which relate primarily to the mean of the distributions of mutational effects and the fraction of slightly deleterious and weakly beneficial mutations that are fixed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
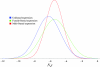
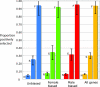


Comment in
-
Profile of Daniel L. Hartl.Proc Natl Acad Sci U S A. 2007 May 29;104(22):9111-3. doi: 10.1073/pnas.0703562104. Epub 2007 May 22. Proc Natl Acad Sci U S A. 2007. PMID: 17519326 Free PMC article. No abstract available.
Similar articles
-
Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection.J Mol Evol. 2003;57 Suppl 1:S154-64. doi: 10.1007/s00239-003-0022-3. J Mol Evol. 2003. PMID: 15008412
-
Inferring the distribution of selective effects from a time inhomogeneous model.PLoS One. 2019 Jan 18;14(1):e0194709. doi: 10.1371/journal.pone.0194709. eCollection 2019. PLoS One. 2019. PMID: 30657757 Free PMC article.
-
The evolution of small insertions and deletions in the coding genes of Drosophila melanogaster.Mol Biol Evol. 2013 Dec;30(12):2699-708. doi: 10.1093/molbev/mst167. Epub 2013 Sep 26. Mol Biol Evol. 2013. PMID: 24077769
-
Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes.BMC Evol Biol. 2008 Dec 18;8:334. doi: 10.1186/1471-2148-8-334. BMC Evol Biol. 2008. PMID: 19091130 Free PMC article.
-
Frequency of spontaneous and induced "point" mutations in higher eukaryotes.J Hered. 1983 Jan-Feb;74(1):2-15. doi: 10.1093/oxfordjournals.jhered.a109711. J Hered. 1983. PMID: 6402543 Review.
Cited by
-
Detection of Regional Variation in Selection Intensity within Protein-Coding Genes Using DNA Sequence Polymorphism and Divergence.Mol Biol Evol. 2017 Nov 1;34(11):3006-3022. doi: 10.1093/molbev/msx213. Mol Biol Evol. 2017. PMID: 28962009 Free PMC article.
-
The stabilization of equilibria in evolutionary game dynamics through mutation: mutation limits in evolutionary games.Proc Math Phys Eng Sci. 2019 Nov;475(2231):20190355. doi: 10.1098/rspa.2019.0355. Epub 2019 Nov 13. Proc Math Phys Eng Sci. 2019. PMID: 31824216 Free PMC article.
-
Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species.Mol Biol Evol. 2009 Jun;26(6):1341-55. doi: 10.1093/molbev/msp043. Epub 2009 Mar 10. Mol Biol Evol. 2009. PMID: 19276154 Free PMC article.
-
Segregating variation in the polycomb group gene cramped alters the effect of temperature on multiple traits.PLoS Genet. 2011 Jan 20;7(1):e1001280. doi: 10.1371/journal.pgen.1001280. PLoS Genet. 2011. PMID: 21283785 Free PMC article.
-
Methods to detect selection on noncoding DNA.Methods Mol Biol. 2012;856:141-59. doi: 10.1007/978-1-61779-585-5_6. Methods Mol Biol. 2012. PMID: 22399458 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

