Identification of polymorphic antioxidant response elements in the human genome
- PMID: 17409198
- PMCID: PMC2805149
- DOI: 10.1093/hmg/ddm066
Identification of polymorphic antioxidant response elements in the human genome
Erratum in
- Hum Mol Genet. 2007 Nov 15;16(22):2780
Abstract
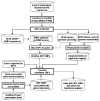
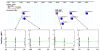
Single nucleotide polymorphisms (SNPs) in transcription factor binding sites (TFBSs) may affect the binding of transcription factors, lead to differences in gene expression and phenotypes and therefore affect susceptibility to environmental exposure. We developed an integrated computational system for discovering functional SNPs in TFBSs in the human genome and predicting their impact on the expression of target genes. In this system, we (i) construct a position weight matrix (PWM) from a collection of experimentally discovered TFBSs; (ii) predict TFBSs in SNP sequences using the PWM and map SNPs to the upstream regions of genes; (iii) examine the evolutionary conservation of putative TFBSs by phylogenetic footprinting; (iv) prioritize candidate SNPs based on microarray expression profiles from tissues in which the transcription factor of interest is either deleted or over-expressed and (v) finally, analyze association of SNP genotypes with gene expression phenotypes. The application of our system has been tested to identify functional polymorphisms in the antioxidant response element (ARE), a cis-acting enhancer sequence found in the promoter region of many genes that encode antioxidant and Phase II detoxification enzymes/proteins. In response to oxidative stress, the transcription factor NRF2 (nuclear factor erythroid-derived 2-like 2) binds to AREs, mediating transcriptional activation of its responsive genes and modulating in vivo defense mechanisms against oxidative damage. Using our novel computational tools, we have identified a set of polymorphic AREs with functional evidence, showing the utility of our system to direct further experimental validation of genomic sequence variations that could be useful for identifying high-risk individuals.
Conflict of interest statement
Figures

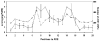

References
-
- Tomso DJ, Bell DA. In: Cellular and Molecular Toxicology. Vanden Heuvel J, editor. Elsevier; 2002. pp. 239–248.
-
- De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. - PubMed
-
- Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995;333:1171–1175. - PubMed
-
- Grant DJ, Hall IJ, Eastmond DA, Jones IM, Bell DA. Bilirubin UDP-glucuronosyltransferase 1A1 (UGT1A1) gene promoter polymorphisms and HPRT, glycophorin A, and micronuclei mutant frequencies in human blood. Mutat Res. 2004;560:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

