DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG)
- PMID: 17410210
- PMCID: PMC1864966
- DOI: 10.1038/sj.emboj.7601649
DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG)
Abstract
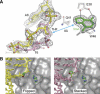
DNA glycosylases help maintain the genome by excising chemically modified bases from DNA. Escherichia coli 3-methyladenine DNA glycosylase I (TAG) specifically catalyzes the removal of the cytotoxic lesion 3-methyladenine (3mA). The molecular basis for the enzymatic recognition and removal of 3mA from DNA is currently a matter of speculation, in part owing to the lack of a structure of a 3mA-specific glycosylase bound to damaged DNA. Here, high-resolution crystal structures of Salmonella typhi TAG in the unliganded form and in a ternary product complex with abasic DNA and 3mA nucleobase are presented. Despite its structural similarity to the helix-hairpin-helix superfamily of DNA glycosylases, TAG has evolved a modified strategy for engaging damaged DNA. In contrast to other glycosylase-DNA structures, the abasic ribose is not flipped into the TAG active site. This is the first structural demonstration that conformational relaxation must occur in the DNA upon base hydrolysis. Together with mutational studies of TAG enzymatic activity, these data provide a model for the specific recognition and hydrolysis of 3mA from DNA.
Figures

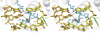
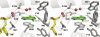


Similar articles
-
Structure of Escherichia coli AlkA in complex with undamaged DNA.J Biol Chem. 2010 Nov 12;285(46):35783-91. doi: 10.1074/jbc.M110.155663. Epub 2010 Sep 15. J Biol Chem. 2010. PMID: 20843803 Free PMC article.
-
Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases.EMBO J. 2003 Oct 1;22(19):4898-909. doi: 10.1093/emboj/cdg505. EMBO J. 2003. PMID: 14517230 Free PMC article.
-
A model for 3-methyladenine recognition by 3-methyladenine DNA glycosylase I (TAG) from Staphylococcus aureus.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Jun 1;68(Pt 6):610-5. doi: 10.1107/S1744309112016363. Epub 2012 May 22. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22684054 Free PMC article.
-
Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase.Mutat Res. 2000 Aug 30;460(3-4):201-10. doi: 10.1016/s0921-8777(00)00027-6. Mutat Res. 2000. PMID: 10946229 Review.
-
DNA glycosylase recognition and catalysis.Curr Opin Struct Biol. 2004 Feb;14(1):43-9. doi: 10.1016/j.sbi.2004.01.003. Curr Opin Struct Biol. 2004. PMID: 15102448 Review.
Cited by
-
Genomic analysis of Oceanotoga teriensis strain UFV_LIMV02, a multidrug-resistant thermophilic bacterium isolated from an offshore oil reservoir.Access Microbiol. 2024 Aug 15;6(8):000801.v3. doi: 10.1099/acmi.0.000801.v3. eCollection 2024. Access Microbiol. 2024. PMID: 39148687 Free PMC article.
-
Characterization of a conserved interaction between DNA glycosylase and ParA in Mycobacterium smegmatis and M. tuberculosis.PLoS One. 2012;7(6):e38276. doi: 10.1371/journal.pone.0038276. Epub 2012 Jun 4. PLoS One. 2012. PMID: 22675536 Free PMC article.
-
The substrate binding interface of alkylpurine DNA glycosylase AlkD.DNA Repair (Amst). 2014 Jan;13:50-4. doi: 10.1016/j.dnarep.2013.10.009. Epub 2013 Nov 26. DNA Repair (Amst). 2014. PMID: 24286669 Free PMC article.
-
Detection of damaged DNA bases by DNA glycosylase enzymes.Biochemistry. 2010 Jun 22;49(24):4957-67. doi: 10.1021/bi100593a. Biochemistry. 2010. PMID: 20469926 Free PMC article. Review.
-
Interplay between base excision repair activity and toxicity of 3-methyladenine DNA glycosylases in an E. coli complementation system.Mutat Res. 2014 May-Jun;763-764:64-73. doi: 10.1016/j.mrfmmm.2014.03.007. Epub 2014 Apr 4. Mutat Res. 2014. PMID: 24709477 Free PMC article.
References
-
- Banerjee A, Santos WL, Verdine GL (2006) Structure of a DNA glycosylase searching for lesions. Science 311: 1153–1157 - PubMed
-
- Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell 92: 117–129 - PubMed
-
- Begley TJ, Haas BJ, Noel J, Shekhtman A, Williams WA, Cunningham RP (1999) A new member of the endonuclease III family of DNA repair enzymes that removes methylated purines from DNA. Curr Biol 9: 653–656 - PubMed
-
- Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E (1994) DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem 269: 30489–30495 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

