Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides
- PMID: 17411013
- PMCID: PMC2538959
- DOI: 10.1021/ac062292d
Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides
Abstract
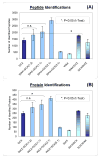
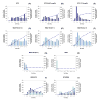
Shotgun proteomics typically uses multidimensional LC/MS/MS analysis of enzymatically digested proteins, where strong cation-exchange (SCX) and reversed-phase (RP) separations are coupled to increase the separation power and dynamic range of analysis. Here we report an on-line multidimensional LC method using an anion- and cation-exchange mixed bed for the first separation dimension. The mixed-bed ion-exchange resin improved peptide recovery over SCX resins alone and showed better orthogonality to RP separations in two-dimensional separations. The Donnan effect, which was enhanced by the introduction of fixed opposite charges in one column, is proposed as the mechanism responsible for improved peptide recovery by producing higher fluxes of salt cations and lower populations of salt anions proximal to the SCX phase. An increase in orthogonality was achieved by a combination of increased retention for acidic peptides and moderately reduced retention of neutral to basic peptides by the added anion-exchange resin. The combination of these effects led to approximately 100% increase in the number of identified peptides from an analysis of a tryptic digest of a yeast whole cell lysate. The application of the method to phosphopeptide-enriched samples increased by 94% phosphopeptide identifications over SCX alone. The lower pKa of phosphopeptides led to specific enrichment in a single salt step resolving acidic phosphopeptides from other phospho- and non-phosphopeptides. Unlike previous methods that use anion exchange to alter selectivity or enrich phosphopeptides, the proposed format is unique in that it works with typical acidic buffer systems used in electrospray ionization, making it feasible for online multidimensional LC/MS/MS applications.
Figures



Similar articles
-
Phosphopeptides enrichment using on-line two-dimensional strong cation exchange followed by reversed-phase liquid chromatography/mass spectrometry.Anal Biochem. 2006 Jul 15;354(2):213-9. doi: 10.1016/j.ab.2006.04.027. Epub 2006 May 11. Anal Biochem. 2006. PMID: 16750159
-
Improving depth in phosphoproteomics by using a strong cation exchange-weak anion exchange-reversed phase multidimensional separation approach.Anal Chem. 2011 Sep 15;83(18):7137-43. doi: 10.1021/ac2015068. Epub 2011 Aug 17. Anal Chem. 2011. PMID: 21815630
-
Fully automatic separation and identification of phosphopeptides by continuous pH-gradient anion exchange online coupled with reversed-phase liquid chromatography mass spectrometry.J Proteome Res. 2009 Jan;8(1):133-41. doi: 10.1021/pr800381w. J Proteome Res. 2009. PMID: 19053533
-
Accessible proteomics space and its implications for peak capacity for zero-, one- and two-dimensional separations coupled with FT-ICR and TOF mass spectrometry.J Mass Spectrom. 2006 Mar;41(3):281-8. doi: 10.1002/jms.1024. J Mass Spectrom. 2006. PMID: 16538648 Review.
-
Multidimensional chromatography coupled to mass spectrometry in analysing complex proteomics samples.J Sep Sci. 2010 Jun;33(10):1421-37. doi: 10.1002/jssc.201000050. J Sep Sci. 2010. PMID: 20486207 Review.
Cited by
-
Proteomic Profiling of Emiliania huxleyi Using a Three-Dimensional Separation Method Combined with Tandem Mass Spectrometry.Molecules. 2020 Jul 2;25(13):3028. doi: 10.3390/molecules25133028. Molecules. 2020. PMID: 32630776 Free PMC article.
-
Online Mixed-Bed Ion Exchange Chromatography for Native Top-Down Proteomics of Complex Mixtures.J Proteome Res. 2024 Jul 5;23(7):2315-2322. doi: 10.1021/acs.jproteome.4c00430. Epub 2024 Jun 24. J Proteome Res. 2024. PMID: 38913967 Free PMC article.
-
Comparative assessment of site assignments in CID and electron transfer dissociation spectra of phosphopeptides discloses limited relocation of phosphate groups.Mol Cell Proteomics. 2010 Oct;9(10):2140-8. doi: 10.1074/mcp.M900619-MCP200. Epub 2010 Mar 16. Mol Cell Proteomics. 2010. PMID: 20233845 Free PMC article.
-
Bifunctional polymer brushes for low-bias enrichment of mono- and multi-phosphorylated peptides prior to mass spectrometry analysis.Analyst. 2011 Sep 21;136(18):3595-8. doi: 10.1039/c1an15489c. Epub 2011 Jul 21. Analyst. 2011. PMID: 21776496 Free PMC article.
-
Hydrophilic interaction liquid chromatography (HILIC) in proteomics.Anal Bioanal Chem. 2008 May;391(1):151-9. doi: 10.1007/s00216-008-1865-7. Epub 2008 Feb 9. Anal Bioanal Chem. 2008. PMID: 18264818 Free PMC article. Review.
References
-
- Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. - PubMed
-
- Wolters DA, Washburn MP, Yates JR., 3rd Anal Chem. 2001;73:5683–5690. - PubMed
-
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Nat Biotechnol. 1999;17:676–682. - PubMed
-
- Giddings JC. Anal Chem. 1984;56:1258A–1260A. 1262A, 1264A. - PubMed
-
- Gilar M, Daly AE, Kele M, Neue UD, Gebler JC. J Chromatogr A. 2004;1061:183–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

