Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver
- PMID: 17412818
- PMCID: PMC2585771
- DOI: 10.1210/en.2006-1192
Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver
Abstract
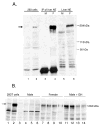
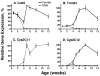
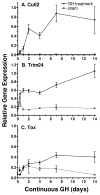
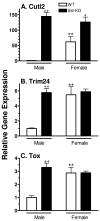
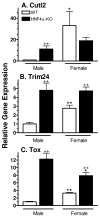
Plasma GH profiles regulate the sexually dimorphic expression of cytochromes P450 and many other genes in rat and mouse liver; however, the proximal transcriptional regulators of these genes are unknown. Presently, we characterize three liver transcription factors that are expressed in adult female rat and mouse liver at levels up to 16-fold [thymus high-mobility group box protein (Tox)], 73-fold [tripartite motif-containing 24 (Trim24)/transcription initiation factor-1alpha (TIF1alpha)], and 125-fold [cut-like 2 (Cutl2)/cut homeobox 2 (Cux2)] higher than in adult males, depending on the strain and species, with Tox expression only detected in mice. In rats, these sex differences first emerged at puberty, when the high prepubertal expression of Cutl2 and Trim24 was extinguished in males but was further increased in females. Rat hepatic expression of Cutl2 and Trim24 was abolished by hypophysectomy and, in the case of Cutl2, was restored to near-female levels by continuous GH replacement. Cutl2 and Trim24 were increased to female-like levels in livers of intact male rats and mice treated with GH continuously (female GH pattern), whereas Tox expression reached only about 40% of adult female levels. Expression of all three genes was also elevated to normal female levels or higher in male mice whose plasma GH profile was feminized secondary to somatostatin gene disruption. Cutl2 and Trim24 both responded to GH infusion in mice within 10-24 h and Tox within 4 d, as compared with at least 4-7 d required for the induced expression of several continuous GH-regulated cytochromes P450 and other female-specific hepatic genes. Cutl2, Trim24, and Tox were substantially up-regulated in livers of male mice deficient in either of two transcription factors implicated in GH regulation of liver sex specificity, namely, signal transducer and activator of transcription 5b (STAT5b) and hepatocyte nuclear factor 4alpha (HNF4alpha), with sex-specific expression being substantially reduced or lost in mice deficient in either nuclear factor. Cutl2 and Trim24 both display transcriptional repressor activity and could thus contribute to the loss of GH-regulated, male-specific liver gene expression seen in male mice deficient in STAT5b or HNF4alpha. Binding sites for Cutl1, whose DNA-binding specificity is close to that of Cutl2, were statistically overrepresented in STAT5b-dependent male-specific mouse genes, lending support to this hypothesis.
Figures



Similar articles
-
Codependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4alpha.Mol Endocrinol. 2006 Mar;20(3):647-60. doi: 10.1210/me.2005-0328. Epub 2005 Oct 20. Mol Endocrinol. 2006. PMID: 16239260
-
Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5.Mol Endocrinol. 2009 Nov;23(11):1914-26. doi: 10.1210/me.2009-0242. Epub 2009 Oct 1. Mol Endocrinol. 2009. PMID: 19797429 Free PMC article.
-
Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2.J Biol Chem. 2005 Feb 4;280(5):3259-68. doi: 10.1074/jbc.M409294200. Epub 2004 Nov 10. J Biol Chem. 2005. PMID: 15539409
-
Role of hepatocyte nuclear factors in growth hormone-regulated, sexually dimorphic expression of liver cytochromes P450.Growth Factors. 2004 Jun;22(2):79-88. doi: 10.1080/08977190410001715172. Growth Factors. 2004. PMID: 15253383 Review.
-
Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression.Novartis Found Symp. 2000;227:61-74; discussion 75-81. doi: 10.1002/0470870796.ch5. Novartis Found Symp. 2000. PMID: 10752065 Review.
Cited by
-
Age and sex dependent changes in liver gene expression during the life cycle of the rat.BMC Genomics. 2010 Nov 30;11:675. doi: 10.1186/1471-2164-11-675. BMC Genomics. 2010. PMID: 21118493 Free PMC article.
-
Sexually dimorphic genome-wide binding of retinoid X receptor alpha (RXRα) determines male-female differences in the expression of hepatic lipid processing genes in mice.PLoS One. 2013 Aug 19;8(8):e71538. doi: 10.1371/journal.pone.0071538. eCollection 2013. PLoS One. 2013. PMID: 23977068 Free PMC article.
-
CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage.J Biol Chem. 2015 Sep 11;290(37):22520-31. doi: 10.1074/jbc.M115.651042. Epub 2015 Jul 28. J Biol Chem. 2015. PMID: 26221032 Free PMC article.
-
Beyond the X Factor: Relevance of Sex Hormones in NAFLD Pathophysiology.Cells. 2021 Sep 21;10(9):2502. doi: 10.3390/cells10092502. Cells. 2021. PMID: 34572151 Free PMC article. Review.
-
Disruption of STAT5b-Regulated Sexual Dimorphism of the Liver Transcriptome by Diverse Factors Is a Common Event.PLoS One. 2016 Mar 9;11(3):e0148308. doi: 10.1371/journal.pone.0148308. eCollection 2016. PLoS One. 2016. PMID: 26959975 Free PMC article.
References
-
- Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Molecular endocrinology (Baltimore, Md. 2006;20:2613–2629. - PubMed
-
- Mode A, Gustafsson JA. Sex and the liver - a journey through five decades. Drug Metab Rev. 2006;38:197–207. - PubMed
-
- Ahluwalia A, Clodfelter KH, Waxman DJ. Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Molecular endocrinology (Baltimore, Md. 2004;18:747–760. - PubMed
-
- Laz EV, Wiwi CA, Waxman DJ. Sexual dimorphism of rat liver nuclear proteins: regulatory role of growth hormone. Mol Cell Proteomics. 2004;3:1170–1180. - PubMed
-
- Eden S. Age- and sex-related differences in episodic growth hormone secretion in the rat. Endocrinology. 1979;105:555–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

