High throughput functional analysis of HIV-1 env genes without cloning
- PMID: 17416428
- PMCID: PMC1948023
- DOI: 10.1016/j.jviromet.2007.02.015
High throughput functional analysis of HIV-1 env genes without cloning
Abstract
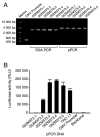
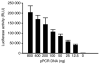
Functional human immunodeficiency virus type 1 (HIV-1) env genes have been widely used for vaccine design, neutralization assays, and pathogenesis studies. However, obtaining bona fide functional env clones is a time consuming and labor intensive process. A new high throughput method has been developed to characterize HIV-1 env genes. Multiple rev/env gene cassettes were obtained from each of seven HIV-1 strains using single genome amplification (SGA) PCR. The cytomegalovirus (CMV) promoter was amplified separately by PCR. A promoter PCR (pPCR) method was developed to link both PCR products using an overlapping PCR method. Pseudovirions were generated by cotransfection of pPCR products and pSG3 Delta env backbone into 293T cells. After infecting TZM-bl cells, 75 out of 87 (86%) of the rev/env gene cassettes were functional. Pseudoviruses generated with pPCR products or corresponding plasmid DNA showed similar sensitivity to six HIV-1 positive sera and three monoclonal antibodies, suggesting neutralization properties are not altered in pPCR pseudovirions. Furthermore, sufficient amounts of pseudovirions can be obtained for a large number of neutralization assays. The new pPCR method eliminates cloning, transformation, and plasmid DNA preparation steps in the generation of HIV-1 pseudovirions. This allows for quick analysis of multiple env genes from HIV-1 infected individuals.
Figures



References
-
- Cordonnier A, Montagnier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989;340:571–4. - PubMed
-
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–22. - PubMed
-
- Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J Virol Methods. 1998;76:139–48. - PubMed
-
- Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996;70:1651–67. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

