Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili
- PMID: 17416646
- PMCID: PMC1913455
- DOI: 10.1128/JB.01942-06
Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili
Erratum in
- J Bacteriol. 2007 Sep;189(17):6507
Abstract
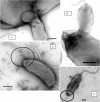
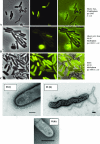
Early electron microscopy and more recent studies in our laboratory of Bdellovibrio bacteriovorus cells indicated the presence of narrow fibers at the nonflagellar pole of this unusual predatory bacterium. Analysis of the B. bacteriovorus HD100 genome showed a complete set of genes potentially encoding type IV pili and an incomplete gene set for Flp pili; therefore, the role of type IV pili in the predatory life cycle of B. bacteriovorus HD100 was investigated. Alignment of the predicted PilA protein with known type IV pilins showed the characteristic conserved N terminus common to type IVa pilins. The pilA gene, encoding the type IV pilus fiber protein, was insertionally inactivated in multiple Bdellovibrio replicate cultures, and the effect upon the expression of other pilus genes was monitored by reverse transcriptase PCR. Interruption of pilA in replicate isolates abolished Bdellovibrio predatory capability in liquid prey cultures and on immobilized yellow fluorescent protein-labeled prey, but the mutants could be cultured prey independently. Expression patterns of pil genes involved in the formation of type IV pili were profiled across the predatory life cycle from attack phase predatory Bdellovibrio throughout the intraperiplasmic bdelloplast stages to prey lysis and in prey-independent growth. Taken together, the data show that type IV pili play a critical role in Bdellovibrio predation.
Figures


References
-
- Barel, G., and E. Jurkevitch. 2001. Analysis of phenotypic diversity among prey-independent mutants of Bdellovibrio bacteriovorus 109J. Arch. Micorbiol. 176:211-216. - PubMed
-
- Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. - PubMed
-
- Burrows, L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878-888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

