Identification of the sigmaB regulon of Bacillus cereus and conservation of sigmaB-regulated genes in low-GC-content gram-positive bacteria
- PMID: 17416654
- PMCID: PMC1913364
- DOI: 10.1128/JB.00313-07
Identification of the sigmaB regulon of Bacillus cereus and conservation of sigmaB-regulated genes in low-GC-content gram-positive bacteria
Abstract
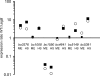
The alternative sigma factor sigma(B) has an important role in the acquisition of stress resistance in many gram-positive bacteria, including the food-borne pathogen Bacillus cereus. Here, we describe the identification of the set of sigma(B)-regulated genes in B. cereus by DNA microarray analysis of the transcriptome upon a mild heat shock. Twenty-four genes could be identified as being sigma(B) dependent as witnessed by (i) significantly lower expression levels of these genes in mutants with a deletion of sigB and rsbY (which encode the alternative sigma factor sigma(B) and a crucial positive regulator of sigma(B) activity, respectively) than in the parental strain B. cereus ATCC 14579 and (ii) increased expression of these genes upon a heat shock. Newly identified sigma(B)-dependent genes in B. cereus include a histidine kinase and two genes that have predicted functions in spore germination. This study shows that the sigma(B) regulon of B. cereus is considerably smaller than that of other gram-positive bacteria. This appears to be in line with phylogenetic analyses where sigma(B) of the B. cereus group was placed close to the ancestral form of sigma(B) in gram-positive bacteria. The data described in this study and previous studies in which the complete sigma(B) regulon of the gram-positive bacteria Bacillus subtilis, Listeria monocytogenes, and Staphylococcus aureus were determined enabled a comparison of the sets of sigma(B)-regulated genes in the different gram-positive bacteria. This showed that only three genes (rsbV, rsbW, and sigB) are conserved in their sigma(B) dependency in all four bacteria, suggesting that the sigma(B) regulon of the different gram-positive bacteria has evolved to perform niche-specific functions.
Figures

Similar articles
-
Deletion of sigB in Bacillus cereus affects spore properties.FEMS Microbiol Lett. 2005 Nov 1;252(1):169-73. doi: 10.1016/j.femsle.2005.08.042. Epub 2005 Sep 9. FEMS Microbiol Lett. 2005. PMID: 16171954
-
The sigmaB regulon in Staphylococcus aureus and its regulation.Int J Med Microbiol. 2006 Aug;296(4-5):237-58. doi: 10.1016/j.ijmm.2005.11.011. Epub 2006 Apr 27. Int J Med Microbiol. 2006. PMID: 16644280
-
The role of sigmaB in the stress response of Gram-positive bacteria -- targets for food preservation and safety.Curr Opin Biotechnol. 2005 Apr;16(2):218-24. doi: 10.1016/j.copbio.2005.01.008. Curr Opin Biotechnol. 2005. PMID: 15831390 Review.
-
A novel hybrid kinase is essential for regulating the sigma(B)-mediated stress response of Bacillus cereus.Environ Microbiol. 2010 Mar;12(3):730-45. doi: 10.1111/j.1462-2920.2009.02116.x. Epub 2009 Dec 3. Environ Microbiol. 2010. PMID: 19958380
-
SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria.Annu Rev Microbiol. 2007;61:215-36. doi: 10.1146/annurev.micro.61.080706.093445. Annu Rev Microbiol. 2007. PMID: 18035607 Review.
Cited by
-
The Alternative Sigma Factor SigB Is Required for the Pathogenicity of Bacillus thuringiensis.J Bacteriol. 2020 Oct 8;202(21):e00265-20. doi: 10.1128/JB.00265-20. Print 2020 Oct 8. J Bacteriol. 2020. PMID: 32817096 Free PMC article.
-
Prediction and validation of novel SigB regulon members in Bacillus subtilis and regulon structure comparison to Bacillales members.BMC Microbiol. 2023 Jan 18;23(1):17. doi: 10.1186/s12866-022-02700-0. BMC Microbiol. 2023. PMID: 36653740 Free PMC article.
-
orf4 of the Bacillus cereus sigB gene cluster encodes a general stress-inducible Dps-like bacterioferritin.J Bacteriol. 2009 Jul;191(14):4522-33. doi: 10.1128/JB.00272-09. Epub 2009 May 8. J Bacteriol. 2009. PMID: 19429618 Free PMC article.
-
The Listeria monocytogenes σB regulon and its virulence-associated functions are inhibited by a small molecule.mBio. 2011 Nov 29;2(6):e00241-11. doi: 10.1128/mBio.00241-11. Print 2011. mBio. 2011. PMID: 22128349 Free PMC article.
-
Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition.Genes Dev. 2009 Oct 15;23(20):2426-36. doi: 10.1101/gad.1843709. Genes Dev. 2009. PMID: 19833768 Free PMC article.
References
-
- den Hengst, C. D., S. A. F. T. van Hijum, J. M. W. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. - PubMed
-
- de Vries, Y. P., L. M. Hornstra, R. D. Atmadja, W. van Schaik, W. M. de Vos, and T. Abee. 2005. Deletion of sigB in Bacillus cereus affects spore properties. FEMS Microbiol. Lett. 252:169-173. - PubMed
-
- Durbin, R., S. R. Eddy, A. Krogh, and G. Mitchison. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

