Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae
- PMID: 17416666
- PMCID: PMC1913403
- DOI: 10.1128/JB.00213-07
Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae
Abstract
The transposon MTnSag1 from Streptococcus agalactiae carried an IS1-like transposase gene and the lnu(C) gene, which encoded a lincosamide nucleotidyltransferase. MTnSag1 could be mobilized by the conjugative transposon Tn916. An intermediate circular form of MTnSag1 and a putative origin of transfer at the 3' end of the lnu(C) gene were characterized.
Figures

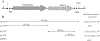

References
-
- Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. - PubMed
-
- Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. - PubMed
-
- Farrow, K. A., D. Lyras, and J. I. Rood. 2001. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 147:2717-2728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

