HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity
- PMID: 17418410
- PMCID: PMC3145406
- DOI: 10.1016/j.ccr.2007.02.006
HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity
Abstract
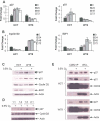
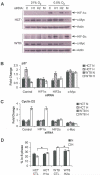
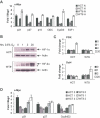
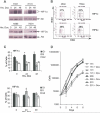
HIF-2alpha promotes von Hippel-Lindau (VHL)-deficient renal clear cell carcinoma (RCC) tumorigenesis, while HIF-1alpha inhibits RCC growth. As HIF-1alpha antagonizes c-Myc function, we hypothesized that HIF-2alpha might enhance c-Myc activity. We demonstrate here that HIF-2alpha promotes cell-cycle progression in hypoxic RCCs and multiple other cell lines. This correlates with enhanced c-Myc promoter binding, transcriptional effects on both activated and repressed target genes, and interactions with Sp1, Miz1, and Max. Finally, HIF-2alpha augments c-Myc transformation of primary mouse embryo fibroblasts (MEFs). Enhanced c-Myc activity likely contributes to HIF-2alpha-mediated neoplastic progression following loss of the VHL tumor suppressor and influences the behavior of hypoxic tumor cells.
Figures




References
-
- Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L, Fukumura D, Moreno-Murciano MP, Herbert JM, Burger A, et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8:131–141. - PubMed
-
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. - PubMed
-
- Baba M, Hirai S, Yamada-Okabe H, Hamada K, Tabuchi H, Kobayashi K, Kondo K, Yoshida M, Yamashita A, Kishida T, et al. Loss of von Hippel-Lindau protein causes cell density dependent deregulation of Cyclin D1 expression through hypoxia-inducible factor. Oncogene. 2003;22:2728–2738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

