Mighty Piwis defend the germline against genome intruders
- PMID: 17418784
- PMCID: PMC4122227
- DOI: 10.1016/j.cell.2007.03.028
Mighty Piwis defend the germline against genome intruders
Abstract
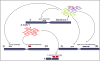
Piwis are a germline-specific subclass of the Argonaute family of RNA interference (RNAi) effector proteins that are associated with a recently discovered group of small RNAs (piRNAs). Recent studies in Drosophila and zebrafish directly implicate Piwi proteins in piRNA biogenesis to maintain transposon silencing in the germline genome (Brennecke et al., 2007; Gunawardane et al., 2007; Houwing et al., 2007). This function may be conserved in mice as loss of Miwi2, a mouse Piwi homolog, leads to germline stem cell and meiotic defects correlated with increased transposon activity (Carmell et al., 2007).
Figures

References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. - PubMed
-
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007:XXX. - PubMed
-
- Bucheton A. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet. 1995;11:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

