Macrocyclic beta-sheet peptides that mimic protein quaternary structure through intermolecular beta-sheet interactions
- PMID: 17419629
- PMCID: PMC2596933
- DOI: 10.1021/ja068511u
Macrocyclic beta-sheet peptides that mimic protein quaternary structure through intermolecular beta-sheet interactions
Abstract
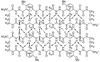
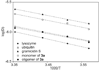
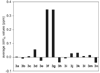
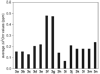
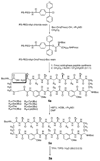
This paper reports the design, synthesis, and characterization of a family of cyclic peptides that mimic protein quaternary structure through beta-sheet interactions. These peptides are 54-membered-ring macrocycles comprising an extended heptapeptide beta-strand, two Hao beta-strand mimics [JACS 2000, 122, 7654] joined by one additional alpha-amino acid, and two delta-linked ornithine beta-turn mimics [JACS 2003, 125, 876]. Peptide 3a, as the representative of these cyclic peptides, contains a heptapeptide sequence (TSFTYTS) adapted from the dimerization interface of protein NuG2 [PDB ID: 1mio]. 1H NMR studies of aqueous solutions of peptide 3a show a partially folded monomer in slow exchange with a strongly folded oligomer. NOE studies clearly show that the peptide self-associates through edge-to-edge beta-sheet dimerization. Pulsed-field gradient (PFG) NMR diffusion coefficient measurements and analytical ultracentrifugation (AUC) studies establish that the oligomer is a tetramer. Collectively, these experiments suggest a model in which cyclic peptide 3a oligomerizes to form a dimer of beta-sheet dimers. In this tetrameric beta-sheet sandwich, the macrocyclic peptide 3a is folded to form a beta-sheet, the beta-sheet is dimerized through edge-to-edge interactions, and this dimer is further dimerized through hydrophobic face-to-face interactions involving the Phe and Tyr groups. Further studies of peptides 3b-3n, which are homologues of peptide 3a with 1-6 variations in the heptapeptide sequence, elucidate the importance of the heptapeptide sequence in the folding and oligomerization of this family of cyclic peptides. Studies of peptides 3b-3g show that aromatic residues across from Hao improve folding of the peptide, while studies of peptides 3h-3n indicate that hydrophobic residues at positions R3 and R5 of the heptapeptide sequence are important in oligomerization.
Figures










References
-
- Maitra S, Nowick JS. In: The Amide Linkage: Structural Significance in Chemistry, Biochemistry, and Materials Science. Greenberg A, Breneman CM, Liebman JF, editors. New York: Wiley; 2000. pp. 495–518.
- Remaut H, Waksman G. Trends Biochem. Sci. 2006;31:436–444. - PubMed
-
- Navia MA, Fitzgerald PMD, McKeever BM, Le CT, Heimbach JC, Herber WK, Sigal IS, Darke PL, Springer JP. Nature. 1989;337:615–620. - PubMed
-
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. Nature. 1995;375:554–560. - PubMed
-
- Ross CA, Poirier MA. Nat. Med. 2004;10 Suppl.:S10–S17. - PubMed
- Tycko R. Curr. Opin. Struct. Biol. 2004;14:96–103. - PubMed
- Makin OS, Serpell LC. FEBS J. 2005;272:5950–5961. - PubMed
- Nelson R, Eisenberg D. Curr. Opin. Struct. Biol. 2006;16:260–265. - PubMed
- Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;75:333–366. - PubMed
- Wetzel R. Acc. Chem. Res. 2006;39:671–679. - PubMed
-
- Sommers WS, Phillips SEV. Nature. 1992;359:387–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

