The three-dimensional structure of human interphase chromosomes is related to the transcriptome map
- PMID: 17420274
- PMCID: PMC1900058
- DOI: 10.1128/MCB.00208-07
The three-dimensional structure of human interphase chromosomes is related to the transcriptome map
Abstract
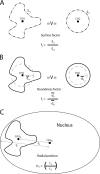
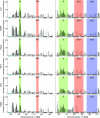
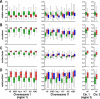
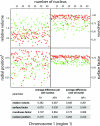
The three-dimensional (3D) organization of the chromosomal fiber in the human interphase nucleus is an important but poorly understood aspect of gene regulation. Here we quantitatively analyze and compare the 3D structures of two types of genomic domains as defined by the human transcriptome map. While ridges are gene dense and show high expression levels, antiridges, on the other hand, are gene poor and carry genes that are expressed at low levels. We show that ridges are in general less condensed, more irregularly shaped, and located more closely to the nuclear center than antiridges. Six human cell lines that display different gene expression patterns and karyotypes share these structural parameters of chromatin. This shows that the chromatin structures of these two types of genomic domains are largely independent of tissue-specific variations in gene expression and differentiation state. Moreover, we show that there is remarkably little intermingling of chromatin from different parts of the same chromosome in a chromosome territory, neither from adjacent nor from distant parts. This suggests that the chromosomal fiber has a compact structure that sterically suppresses intermingling. Together, our results reveal novel general aspects of 3D chromosome architecture that are related to genome structure and function.
Figures



References
-
- Archidiacono, N., R. Antonacci, R. Marzella, P. Finelli, A. Lonoce, and M. Rocchi. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics 25:477-484. - PubMed
-
- Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis, and W. A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211-219. - PubMed
-
- Bridger, J. M., S. Boyle, I. R. Kill, and W. A. Bickmore. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 10:149-152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
