Direct and adaptor-mediated substrate recognition by an essential AAA+ protease
- PMID: 17420450
- PMCID: PMC1871830
- DOI: 10.1073/pnas.0701776104
Direct and adaptor-mediated substrate recognition by an essential AAA+ protease
Abstract
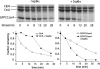
Regulated proteolysis is required to execute many cellular programs. In Caulobacter crescentus, timely degradation of the master regulator CtrA by ClpXP protease is essential for cell-cycle progression and requires the colocalization of CtrA and RcdA. Here, we establish a biochemical framework to understand regulated proteolysis in C. crescentus and show that RcdA is not an adaptor for CtrA degradation. CtrA is rapidly degraded without RcdA and is recognized with an affinity comparable with the best ClpXP substrates. In contrast, SspBalpha, the alpha-proteobacterial homolog of SspB, functions as an adaptor to enhance degradation of specific substrates. Cargo-free SspBalpha is also itself a substrate of ClpXP-mediated proteolysis. Thus, our analysis (i) reveals the consequences of both direct and adaptor-stimulated recognition in mediating substrate specificity in vitro, (ii) reveals a potential regulatory role of controlled adaptor stability, and (iii) suggests that cell-cycle regulation of CtrA stability depends on repression of its intrinsic degradation rather than adaptor-mediated enhancement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

