Structural and chemical profiling of the human cytosolic sulfotransferases
- PMID: 17425406
- PMCID: PMC1847840
- DOI: 10.1371/journal.pbio.0050097
Structural and chemical profiling of the human cytosolic sulfotransferases
Erratum in
- PLoS Biol. 2007 Jun;5(6):e165. Thonton, Janet [corrected to Thornton, Janet]
Abstract
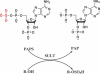
The human cytosolic sulfotransfases (hSULTs) comprise a family of 12 phase II enzymes involved in the metabolism of drugs and hormones, the bioactivation of carcinogens, and the detoxification of xenobiotics. Knowledge of the structural and mechanistic basis of substrate specificity and activity is crucial for understanding steroid and hormone metabolism, drug sensitivity, pharmacogenomics, and response to environmental toxins. We have determined the crystal structures of five hSULTs for which structural information was lacking, and screened nine of the 12 hSULTs for binding and activity toward a panel of potential substrates and inhibitors, revealing unique "chemical fingerprints" for each protein. The family-wide analysis of the screening and structural data provides a comprehensive, high-level view of the determinants of substrate binding, the mechanisms of inhibition by substrates and environmental toxins, and the functions of the orphan family members SULT1C3 and SULT4A1. Evidence is provided for structural "priming" of the enzyme active site by cofactor binding, which influences the spectrum of small molecules that can bind to each enzyme. The data help explain substrate promiscuity in this family and, at the same time, reveal new similarities between hSULT family members that were previously unrecognized by sequence or structure comparison alone.
Conflict of interest statement
Figures







References
-
- Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129:141–170. - PubMed
-
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. - PubMed
-
- Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: Identification of seven novel genes and pseudogenes. Pharmacogenomics J. 2004;4:54–65. - PubMed
-
- Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, et al. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

