Modelling the evolution of the archeal tryptophan synthase
- PMID: 17425797
- PMCID: PMC1854888
- DOI: 10.1186/1471-2148-7-59
Modelling the evolution of the archeal tryptophan synthase
Abstract
Background: Microorganisms and plants are able to produce tryptophan. Enzymes catalysing the last seven steps of tryptophan biosynthesis are encoded in the canonical trp operon. Among the trp genes are most frequently trpA and trpB, which code for the alpha and beta subunit of tryptophan synthase. In several prokaryotic genomes, two variants of trpB (named trpB1 or trpB2) occur in different combinations. The evolutionary history of these trpB genes is under debate.
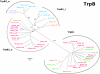
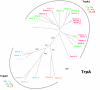
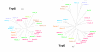
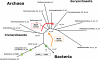
Results: In order to study the evolution of trp genes, completely sequenced archeal and bacterial genomes containing trpB were analysed. Phylogenetic trees indicated that TrpB sequences constitute four distinct groups; their composition is in agreement with the location of respective genes. The first group consisted exclusively of trpB1 genes most of which belonged to trp operons. Groups two to four contained trpB2 genes. The largest group (trpB2_o) contained trpB2 genes all located outside of operons. Most of these genes originated from species possessing an operon-based trpB1 in addition. Groups three and four pertain to trpB2 genes of those genomes containing exclusively one or two trpB2 genes, but no trpB1. One group (trpB2_i) consisted of trpB2 genes located inside, the other (trpB2_a) of trpB2 genes located outside the trp operon. TrpA and TrpB form a heterodimer and cooperate biochemically. In order to characterise trpB variants and stages of TrpA/TrpB cooperation in silico, several approaches were combined. Phylogenetic trees were constructed for all trp genes; their structure was assessed via bootstrapping. Alternative models of trpB evolution were evaluated with parsimony arguments. The four groups of trpB variants were correlated with archeal speciation. Several stages of TrpA/TrpB cooperation were identified and trpB variants were characterised. Most plausibly, trpB2 represents the predecessor of the modern trpB gene, and trpB1 evolved in an ancestral bacterium.
Conclusion: In archeal genomes, several stages of trpB evolution, TrpA/TrpB cooperation, and operon formation can be observed. Thus, archeal trp genes may serve as a model system for studying the evolution of protein-protein interactions and operon formation.
Figures






Similar articles
-
The tryptophan synthase β-subunit paralogs TrpB1 and TrpB2 in Thermococcus kodakarensis are both involved in tryptophan biosynthesis and indole salvage.FEBS J. 2014 Jul;281(14):3113-25. doi: 10.1111/febs.12845. Epub 2014 Jun 10. FEBS J. 2014. PMID: 24835339
-
Evolution of multi-enzyme complexes: the case of tryptophan synthase.Biochemistry. 2006 Nov 28;45(47):14111-9. doi: 10.1021/bi061684b. Biochemistry. 2006. PMID: 17115706
-
A novel tryptophan synthase beta-subunit from the hyperthermophile Thermotoga maritima. Quaternary structure, steady-state kinetics, and putative physiological role.J Biol Chem. 2002 Mar 8;277(10):8194-201. doi: 10.1074/jbc.M111541200. Epub 2001 Dec 26. J Biol Chem. 2002. PMID: 11756459
-
Ancient origin of the tryptophan operon and the dynamics of evolutionary change.Microbiol Mol Biol Rev. 2003 Sep;67(3):303-42, table of contents. doi: 10.1128/MMBR.67.3.303-342.2003. Microbiol Mol Biol Rev. 2003. PMID: 12966138 Free PMC article. Review.
-
Structure and function of archaeal RNA polymerases.Mol Microbiol. 2007 Sep;65(6):1395-404. doi: 10.1111/j.1365-2958.2007.05876.x. Epub 2007 Aug 14. Mol Microbiol. 2007. PMID: 17697097 Review.
Cited by
-
TRAP-5' stem loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region.RNA. 2007 Nov;13(11):2020-33. doi: 10.1261/rna.719507. Epub 2007 Sep 19. RNA. 2007. PMID: 17881743 Free PMC article.
-
TrpY regulation of trpB2 transcription in Methanothermobacter thermautotrophicus.J Bacteriol. 2008 Apr;190(7):2637-41. doi: 10.1128/JB.01926-07. Epub 2008 Feb 8. J Bacteriol. 2008. PMID: 18263726 Free PMC article.
-
Tryptophan Operon Diversity Reveals Evolutionary Trends among Geographically Disparate Chlamydia trachomatis Ocular and Urogenital Strains Affecting Tryptophan Repressor and Synthase Function.mBio. 2021 May 11;12(3):e00605-21. doi: 10.1128/mBio.00605-21. mBio. 2021. PMID: 33975934 Free PMC article.
-
The tryptophan pathway genes of the Sargasso Sea metagenome: new operon structures and the prevalence of non-operon organization.Genome Biol. 2008 Jan 27;9(1):R20. doi: 10.1186/gb-2008-9-1-r20. Genome Biol. 2008. PMID: 18221558 Free PMC article.
-
Evolution of bacterial trp operons and their regulation.Curr Opin Microbiol. 2008 Apr;11(2):78-86. doi: 10.1016/j.mib.2008.02.005. Curr Opin Microbiol. 2008. PMID: 18374625 Free PMC article. Review.
References
-
- Yanofsky C. Using studies on tryptophan metabolism to answer basic biological questions. J Biol Chem. 2003;278:10859–10878. - PubMed
-
- Yanofsky C. Advancing our knowledge in biochemistry, genetics, and microbiology through studies on tryptophan metabolism. Annu Rev Biochem. 2001;70:1–37. - PubMed
-
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. - PubMed
-
- Yanofsky C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004;20:367–374. - PubMed
-
- Gutierrez-Preciado A, Jensen RA, Yanofsky C, Merino E. New insights into regulation of the tryptophan biosynthetic operon in Gram-positive bacteria. Trends Genet. 2005;21:432–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

