DNA sequencing by MALDI-TOF MS using alkali cleavage of RNA/DNA chimeras
- PMID: 17426131
- PMCID: PMC1885642
- DOI: 10.1093/nar/gkm056
DNA sequencing by MALDI-TOF MS using alkali cleavage of RNA/DNA chimeras
Abstract
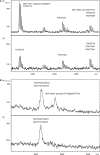
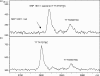
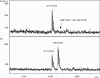
Approaches developed for sequencing DNA with detection by mass spectrometry use strategies that deviate from the Sanger-type methods. Procedures demonstrated so far used the sequence specificity of RNA endonucleases, as unfortunately equivalent enzymes for DNA do not exist and therefore require transcription of DNA into RNA prior to fragmentation. We have developed a novel, rapid and accurate concept for DNA sequencing using mass spectrometry and RNA/DNA chimeras and applied it to sequence mitochondrial DNA. Our method is based on the preparation of a chimeric RNA/DNA with a DNA polymerase that also incorporates ribonucleotides. Sequencing is carried out with one ribonucleotide (ATP, CTP or GTP) and the other three nucleotides in their deoxyribo-form. The product is treated with alkali, which cleaves 3' of all ribonucleotides to form a terminal 3' phosphate. Conditions have been streamlined so that molecular, biological and alkali cleavage conditions are compatible with matrix-assisted laser desorption/ionization time-of-flight (MALDI) mass spectrometric analysis. Fragment analysis by MALDI MS provides a sequence-specific fingerprint, which allows the identification of differences between a reference and another sequence. Due to the mass profile, the position and kind of the mutation can be assigned. These differences between signatures are indicative of known, unidentified, rare and private mutations. This novel DNA sequencing protocol was applied to sequence the hypervariable region 1 (HV1) of mitochondrial DNA in 22 individuals.
Figures

Similar articles
-
High-specificity single-tube multiplex genotyping using Ribo-PAP PCR, tag primers, alkali cleavage of RNA/DNA chimeras and MALDI-TOF MS.Hum Mutat. 2013 Jan;34(1):266-73. doi: 10.1002/humu.22227. Epub 2012 Nov 8. Hum Mutat. 2013. PMID: 23132774
-
SNP genotyping using alkali cleavage of RNA/DNA chimeras and MALDI time-of-flight mass spectrometry.Nucleic Acids Res. 2006 Feb 9;34(3):e18. doi: 10.1093/nar/gnj021. Nucleic Acids Res. 2006. PMID: 16473841 Free PMC article.
-
A revisit of high collision energy effects on collision-induced dissociation spectra using matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-LIFT-TOF/TOF): application to the sequencing of RNA/DNA chimeras.Rapid Commun Mass Spectrom. 2014 Jul 15;28(13):1433-43. doi: 10.1002/rcm.6913. Rapid Commun Mass Spectrom. 2014. PMID: 24861592
-
Mass-spectrometry DNA sequencing.Mutat Res. 2005 Jun 3;573(1-2):3-12. doi: 10.1016/j.mrfmmm.2004.07.021. Mutat Res. 2005. PMID: 15829234 Review.
-
MALDI-TOF-MS analysis of protein and DNA.Neuroscientist. 2001 Feb;7(1):6-12. doi: 10.1177/107385840100700104. Neuroscientist. 2001. PMID: 11486345 Review.
Cited by
-
Limited reverse transcriptase activity of phi29 DNA polymerase.Nucleic Acids Res. 2018 Apr 20;46(7):3625-3632. doi: 10.1093/nar/gky190. Nucleic Acids Res. 2018. PMID: 29554297 Free PMC article.
-
Alkaline hydrolysis to remove potentially infectious viral RNA contaminants from DNA.Virol J. 2016 Jun 4;13:88. doi: 10.1186/s12985-016-0552-0. Virol J. 2016. PMID: 27260412 Free PMC article.
-
Using surface-assisted laser desorption/ionization mass spectrometry to detect ss- and ds-oligodeoxynucleotides.J Am Soc Mass Spectrom. 2013 Jun;24(6):877-83. doi: 10.1007/s13361-013-0595-z. Epub 2013 Mar 29. J Am Soc Mass Spectrom. 2013. PMID: 23539494
References
-
- Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. - PubMed
-
- Nordhoff E, Kirpekar F, Roepstorff P. Mass spectrometry of nucleic acids. Mass Spectrom. Rev. 1996;15:67–138. - PubMed
-
- Tost J, Gut IG. Genotyping single nucleotide polymorphisms by mass spectrometry. Mass Spectrom. Rev. 2002;21:388–418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

