Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function
- PMID: 17426288
- PMCID: PMC2075475
- DOI: 10.1210/me.2006-0389
Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function
Abstract
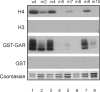
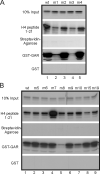
Protein arginine methyltransferase 1 (PRMT1) is an arginine-specific protein methyltransferase that methylates a number of proteins involved in transcription and other aspects of RNA metabolism. Its role as a transcriptional coactivator for nuclear receptors involves its ability to bind to other coactivators, such as glucocorticoid receptor-interacting protein 1 (GRIP1), as well as its ability to methylate histone H4 and coactivators such as peroxisome proliferator-activated receptor gamma coactivator-1alpha. Its ability to form homodimers or higher-order homo-oligomers also is important for its methyltransferase activity. To understand the function of PRMT1 further, 19 surface residues were mutated, based on the crystal structure of PRMT1. Mutants were characterized for their ability to bind and methylate various substrates, form homodimers, bind GRIP1, and function as a coactivator for the androgen receptor in cooperation with GRIP1. We identified specific surface residues that are important for methylation substrate specificity and binding of substrates, for dimerization/oligomerization, and for coactivator function. This analysis also revealed functional relationships between the various activities of PRMT1. Mutants that did not dimerize well had poor methyltransferase activity and coactivator function. However, surprisingly, all dimerization mutants exhibited increased GRIP1 binding, suggesting that the essential PRMT1 coactivator function of binding to GRIP1 may require dissociation of PRMT1 dimers or oligomers. Three different mutants with altered substrate specificity had widely varying coactivator activity levels, suggesting that methylation of specific substrates is important for coactivator function. Finally, identification of several mutants that exhibited reduced coactivator function but appeared normal in all other activities tested, and finding one mutant with very little methyltransferase activity but normal coactivator function, suggested that these mutated surface residues may be involved in currently unknown protein-protein interactions that are important for coactivator function.
Figures





References
-
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. - PubMed
-
- Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. - PubMed
-
- McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. - PubMed
-
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

