Human GLI3 intragenic conserved non-coding sequences are tissue-specific enhancers
- PMID: 17426814
- PMCID: PMC1838922
- DOI: 10.1371/journal.pone.0000366
Human GLI3 intragenic conserved non-coding sequences are tissue-specific enhancers
Abstract
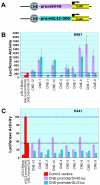
The zinc-finger transcription factor GLI3 is a key regulator of development, acting as a primary transducer of Sonic hedgehog (SHH) signaling in a combinatorial context dependent fashion controlling multiple patterning steps in different tissues/organs. A tight temporal and spatial control of gene expression is indispensable, however, cis-acting sequence elements regulating GLI3 expression have not yet been reported. We show that 11 ancient genomic DNA signatures, conserved from the pufferfish Takifugu (Fugu) rubripes to man, are distributed throughout the introns of human GLI3. They map within larger conserved non-coding elements (CNEs) that are found in the tetrapod lineage. Full length CNEs transiently transfected into human cell cultures acted as cell type specific enhancers of gene transcription. The regulatory potential of these elements is conserved and was exploited to direct tissue specific expression of a reporter gene in zebrafish embryos. Assays of deletion constructs revealed that the human-Fugu conserved sequences within the GLI3 intronic CNEs were essential but not sufficient for full-scale transcriptional activation. The enhancer activity of the CNEs is determined by a combinatorial effect of a core sequence conserved between human and teleosts (Fugu) and flanking tetrapod-specific sequences, suggesting that successive clustering of sequences with regulatory potential around an ancient, highly conserved nucleus might be a possible mechanism for the evolution of cis-acting regulatory elements.
Conflict of interest statement
Figures






References
-
- Radhakrishna U, Bornholdt D, Scott HS, Patel UC, Rossier C, et al. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; No phenotype prediction from the position of GLI3 mutations. Am J Hum Genet. 1999;65:645–655. - PMC - PubMed
-
- Vortkamp A, Gessler M, Grzeschik KH. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352:539–540. - PubMed
-
- Wild A, Kalff-Suske M, Vortkamp A, Bornholdt D, Konig R, et al. Point mutations in human GLI3 cause Greig syndrome. Hum Mol Genet. 1997;6:1979–1984. - PubMed
-
- Kalff-Suske M, Wild A, Topp J, Wessling M, Jacobsen EM, et al. Point mutations throughout the GLI3 gene cause Greig cephalopolysyndactyly syndrome. Hum Mol Genet. 1999;8:1769–1777. - PubMed
-
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

