Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer
- PMID: 17428314
- PMCID: PMC1868918
- DOI: 10.1186/bcr1667
Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer
Abstract
Introduction: The impact of interactions between the two estrogen receptor (ER) subtypes, ERalpha and ERbeta, on gene expression in breast cancer biology is not clear. The goal of this study was to examine transcriptomic alterations in cancer cells co-expressing both receptors and the association of gene expression signatures with disease outcome.
Methods: Transcriptional effects of ERbeta overexpression were determined in a stably transfected cell line derived from ERalpha-positive T-47D cells. Microarray analysis was carried out to identify differential gene expression in the cell line, and expression of key genes was validated by quantitative polymerase chain reaction. Microarray and clinical data from patient samples were then assessed to determine the in vivo relevance of the expression profiles observed in the cell line.
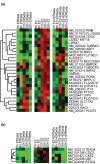
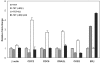
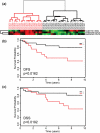
Results: A subset of 14 DNA replication and cell cycle-related genes was found to be specifically downregulated by ERbeta. Expression profiles of four genes, CDC2, CDC6, CKS2, and DNA2L, were significantly inversely correlated with ERbeta transcript levels in patient samples, consistent with in vitro observations. Kaplan-Meier analysis revealed better disease outcome for the patient group with an expression signature linked to higher ERbeta expression as compared to the lower ERbeta-expressing group for both disease-free survival (p = 0.00165) and disease-specific survival (p = 0.0268). These findings were further validated in an independent cohort.
Conclusion: Our findings revealed a transcriptionally regulated mechanism for the previously described growth inhibitory effects of ERbeta in ERalpha-positive breast tumor cells and provide evidence for a functional and beneficial impact of ERbeta in primary breast tumors.
Figures

Comment in
-
Estrogen receptor-beta: why may it influence clinical outcome in estrogen receptor-alpha positive breast cancer?Breast Cancer Res. 2007;9(3):107. doi: 10.1186/bcr1735. Breast Cancer Res. 2007. PMID: 17617929 Free PMC article.
Similar articles
-
ERbeta sensitizes breast cancer cells to retinoic acid: evidence of transcriptional crosstalk.Mol Cancer Res. 2004 Sep;2(9):523-31. Mol Cancer Res. 2004. PMID: 15383631
-
Estrogen receptor beta expression is associated with tamoxifen response in ERalpha-negative breast carcinoma.Clin Cancer Res. 2007 Apr 1;13(7):1987-94. doi: 10.1158/1078-0432.CCR-06-1823. Clin Cancer Res. 2007. PMID: 17404078
-
Molecular identification of ERalpha-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes.J Cell Physiol. 2004 Sep;200(3):440-50. doi: 10.1002/jcp.20039. J Cell Physiol. 2004. PMID: 15254972
-
Clinical significance of estrogen receptor beta in breast cancer.Cancer Chemother Pharmacol. 2005 Nov;56 Suppl 1:21-6. doi: 10.1007/s00280-005-0107-3. Cancer Chemother Pharmacol. 2005. PMID: 16273360 Review.
-
[Estrogen receptor Beta isoforms -- functions and clinical relevance in breast cancer].Zentralbl Gynakol. 2005 Aug;127(4):228-34. doi: 10.1055/s-2005-836563. Zentralbl Gynakol. 2005. PMID: 16037904 Review. German.
Cited by
-
Estrogen Signaling and Its Potential as a Target for Therapy in Ovarian Cancer.Cancers (Basel). 2020 Jun 22;12(6):1647. doi: 10.3390/cancers12061647. Cancers (Basel). 2020. PMID: 32580290 Free PMC article. Review.
-
ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches.Biochim Biophys Acta. 2009 Dec;1796(2):162-75. doi: 10.1016/j.bbcan.2009.06.003. Epub 2009 Jun 13. Biochim Biophys Acta. 2009. PMID: 19527773 Free PMC article. Review.
-
Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis.Genes Dis. 2018 May 12;5(2):77-106. doi: 10.1016/j.gendis.2018.05.001. eCollection 2018 Jun. Genes Dis. 2018. PMID: 30258937 Free PMC article. Review.
-
ERbeta in breast cancer--onlooker, passive player, or active protector?Steroids. 2008 Oct;73(11):1039-51. doi: 10.1016/j.steroids.2008.04.006. Epub 2008 Apr 20. Steroids. 2008. PMID: 18501937 Free PMC article. Review.
-
Integrative genomics of gene and metabolic regulation by estrogen receptors α and β, and their coregulators.Mol Syst Biol. 2013 Jun 18;9:676. doi: 10.1038/msb.2013.28. Mol Syst Biol. 2013. PMID: 23774759 Free PMC article.
References
-
- Parker MG. Structure and function of estrogen receptors. Vitam Horm. 1995;51:267–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

