Comparative genomics using Fugu reveals insights into regulatory subfunctionalization
- PMID: 17428329
- PMCID: PMC1896008
- DOI: 10.1186/gb-2007-8-4-r53
Comparative genomics using Fugu reveals insights into regulatory subfunctionalization
Abstract
Background: A major mechanism for the preservation of gene duplicates in the genome is thought to be mediated via loss or modification of cis-regulatory subfunctions between paralogs following duplication (a process known as regulatory subfunctionalization). Despite a number of gene expression studies that support this mechanism, no comprehensive analysis of regulatory subfunctionalization has been undertaken at the level of the distal cis-regulatory modules involved. We have exploited fish-mammal genomic alignments to identify and compare more than 800 conserved non-coding elements (CNEs) that associate with genes that have undergone fish-specific duplication and retention.
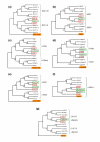
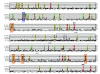
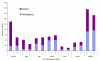
Results: Using the abundance of duplicated genes within the Fugu genome, we selected seven pairs of teleost-specific paralogs involved in early vertebrate development, each containing clusters of CNEs in their vicinity. CNEs present around each Fugu duplicated gene were identified using multiple alignments of orthologous regions between single-copy mammalian orthologs (representing the ancestral locus) and each fish duplicated region in turn. Comparative analysis reveals a pattern of element retention and loss between paralogs indicative of subfunctionalization, the extent of which differs between duplicate pairs. In addition to complete loss of specific regulatory elements, a number of CNEs have been retained in both regions but may be responsible for more subtle levels of subfunctionalization through sequence divergence.
Conclusion: Comparative analysis of conserved elements between duplicated genes provides a powerful approach for studying regulatory subfunctionalization at the level of the regulatory elements involved.
Figures



Similar articles
-
Comparative genomics using fugu: a tool for the identification of conserved vertebrate cis-regulatory elements.Bioessays. 2005 Jan;27(1):100-7. doi: 10.1002/bies.20134. Bioessays. 2005. PMID: 15612032 Review.
-
Analyses of fugu hoxa2 genes provide evidence for subfunctionalization of neural crest cell and rhombomere cis-regulatory modules during vertebrate evolution.Dev Biol. 2016 Jan 15;409(2):530-42. doi: 10.1016/j.ydbio.2015.11.006. Epub 2015 Nov 26. Dev Biol. 2016. PMID: 26632170
-
Ancient duplicated conserved noncoding elements in vertebrates: a genomic and functional analysis.Genome Res. 2006 Apr;16(4):451-65. doi: 10.1101/gr.4143406. Epub 2006 Mar 13. Genome Res. 2006. PMID: 16533910 Free PMC article.
-
CONDOR: a database resource of developmentally associated conserved non-coding elements.BMC Dev Biol. 2007 Aug 30;7:100. doi: 10.1186/1471-213X-7-100. BMC Dev Biol. 2007. PMID: 17760977 Free PMC article.
-
Evolution of the duplicated intracellular lipid-binding protein genes of teleost fishes.Mol Genet Genomics. 2017 Aug;292(4):699-727. doi: 10.1007/s00438-017-1313-5. Epub 2017 Apr 7. Mol Genet Genomics. 2017. PMID: 28389698 Review.
Cited by
-
Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids.Plant Physiol. 2008 Dec;148(4):1772-81. doi: 10.1104/pp.108.124867. Epub 2008 Oct 24. Plant Physiol. 2008. PMID: 18952863 Free PMC article.
-
Efficient large-scale protein sequence comparison and gene matching to identify orthologs and co-orthologs.Nucleic Acids Res. 2012 Mar;40(6):e44. doi: 10.1093/nar/gkr1261. Epub 2011 Dec 30. Nucleic Acids Res. 2012. PMID: 22210858 Free PMC article.
-
Deeply conserved chordate noncoding sequences preserve genome synteny but do not drive gene duplicate retention.Genome Res. 2009 Nov;19(11):2036-51. doi: 10.1101/gr.093237.109. Epub 2009 Aug 24. Genome Res. 2009. PMID: 19704032 Free PMC article.
-
Identification of two evolutionarily conserved 5' cis-elements involved in regulating spatiotemporal expression of Nolz-1 during mouse embryogenesis.PLoS One. 2013;8(1):e54485. doi: 10.1371/journal.pone.0054485. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23349903 Free PMC article.
-
Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence.PLoS Genet. 2008 Feb;4(2):e29. doi: 10.1371/journal.pgen.0040029. PLoS Genet. 2008. PMID: 18282108 Free PMC article.
References
-
- Ohno S. Evolution by Gene Duplication. Heidelberg: Springer-Verlag; 1970.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

