RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions
- PMID: 17428802
- PMCID: PMC2954279
- DOI: 10.1074/jbc.M611392200
RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions
Abstract
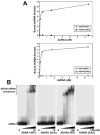
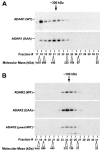
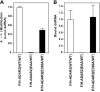
RNA editing that converts adenosine to inosine in double-stranded RNA (dsRNA) is mediated by adenosine deaminases acting on RNA (ADAR). ADAR1 and ADAR2 form respective homodimers, and this association is essential for their enzymatic activities. In this investigation, we set out experiments aiming to determine whether formation of the homodimer complex is mediated by an amino acid interface made through protein-protein interactions of two monomers or via binding of the two subunits to a dsRNA substrate. Point mutations were created in the dsRNA binding domains (dsRBDs) that abolished all RNA binding, as tested for two classes of ADAR ligands, long and short dsRNA. The mutant ADAR dimer complexes were intact, as demonstrated by their ability to co-purify in a sequential affinity-tagged purification and also by their elution at the dimeric fraction position on a size fractionation column. Our results demonstrated ADAR dimerization independent of their binding to dsRNA, establishing the importance of protein-protein interactions for dimer formation. As expected, these mutant ADARs could no longer perform their catalytic function due to the loss in substrate binding. Surprisingly, a chimeric dimer consisting of one RNA binding mutant monomer and a wild type partner still abolished its ability to bind and edit its substrate, indicating that ADAR dimers require two subunits with functional dsRBDs for binding to a dsRNA substrate and then for editing activity to occur.
Figures



References
-
- DeCerbo J, Carmichael GG. Curr Opin Cell Biol. 2005;17:302–308. - PubMed
-
- Reenan RA. Trends Genet. 2001;17:53–56. - PubMed
-
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. - PubMed
-
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. Cell. 1993;75:1361–1370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

