Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing
- PMID: 17428870
- PMCID: PMC1933295
- DOI: 10.1128/JVI.00049-07
Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing
Abstract
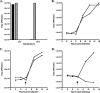
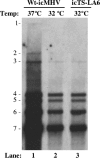
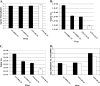
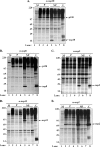
Coronaviruses are the largest RNA viruses, and their genomes encode replication machinery capable of efficient replication of both positive- and negative-strand viral RNAs as well as enzymes capable of processing large viral polyproteins into putative replication intermediates and mature proteins. A model described recently by Sawicki et al. (S. G. Sawicki, D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell, PLoS Pathog. 1:e39, 2005), based upon complementation studies of known temperature-sensitive (TS) mutants of murine hepatitis virus (MHV) strain A59, proposes that an intermediate comprised of nsp4 to nsp10/11 ( approximately 150 kDa) is involved in negative-strand synthesis. Furthermore, the mature forms of nsp4 to nsp10 are thought to serve as cofactors with other replicase proteins to assemble a larger replication complex specifically formed to transcribe positive-strand RNAs. In this study, we introduced a single-amino-acid change (nsp10:Q65E) associated with the TS-LA6 phenotype into nsp10 of the infectious clone of MHV. Growth kinetic studies demonstrated that this mutation was sufficient to generate the TS phenotype at permissive and nonpermissive temperatures. Our results demonstrate that the TS mutant variant of nsp10 inhibits the main protease, 3CLpro, blocking its function completely at the nonpermissive temperature. These results implicate nsp10 as being a critical factor in the activation of 3CLpro function. We discuss how these findings challenge the current hypothesis that nsp4 to nsp10/11 functions as a single cistron in negative-strand RNA synthesis and analyze recent complementation data in light of these new findings.
Figures




Similar articles
-
Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication.J Virol. 2007 Oct;81(19):10280-91. doi: 10.1128/JVI.00017-07. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634238 Free PMC article.
-
A new cistron in the murine hepatitis virus replicase gene.J Virol. 2010 Oct;84(19):10148-58. doi: 10.1128/JVI.00901-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668085 Free PMC article.
-
Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis.J Virol. 2007 Jun;81(12):6356-68. doi: 10.1128/JVI.02805-06. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392363 Free PMC article.
-
Functional and genetic analysis of coronavirus replicase-transcriptase proteins.PLoS Pathog. 2005 Dec;1(4):e39. doi: 10.1371/journal.ppat.0010039. Epub 2005 Dec 9. PLoS Pathog. 2005. PMID: 16341254 Free PMC article. Review.
-
Polyprotein synthesis: a journey from the traditional pre-translational method to modern post-translational approaches for single-molecule force spectroscopy.Chem Commun (Camb). 2023 Jun 6;59(46):6946-6955. doi: 10.1039/d3cc01756g. Chem Commun (Camb). 2023. PMID: 37183922 Review.
Cited by
-
Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity.J Virol. 2012 May;86(9):4801-10. doi: 10.1128/JVI.06754-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345451 Free PMC article.
-
Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase.J Virol. 2010 Jul;84(14):7325-36. doi: 10.1128/JVI.02490-09. Epub 2010 May 5. J Virol. 2010. PMID: 20444893 Free PMC article.
-
Characterization of Bafinivirus main protease autoprocessing activities.J Virol. 2011 Feb;85(3):1348-59. doi: 10.1128/JVI.01716-10. Epub 2010 Nov 10. J Virol. 2011. PMID: 21068254 Free PMC article.
-
Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation.Microbiol Mol Biol Rev. 2008 Dec;72(4):672-85, Table of Contents. doi: 10.1128/MMBR.00015-08. Microbiol Mol Biol Rev. 2008. PMID: 19052324 Free PMC article. Review.
-
Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication.J Virol. 2007 Oct;81(19):10280-91. doi: 10.1128/JVI.00017-07. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634238 Free PMC article.
References
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Baker, S. C. 2004. Coronaviruses: from common colds to severe acute respiratory syndrome. Pediatr. Infect. Dis. J. 23:1049-1050. - PubMed
-
- Baker, S. C., H. Gao, and R. S. Baric. 1993. Altered proteolytic processing of the polymerase polyprotein in RNA (−) temperature sensitive mutants of murine coronavirus. Adv. Exp. Med. Biol. 342:215-219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI023946/AI/NIAID NIH HHS/United States
- DK58404/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- EY08126/EY/NEI NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- HD15052/HD/NICHD NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
- R01 AI023946/AI/NIAID NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- DK59637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources

