Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies
- PMID: 17432829
- PMCID: PMC2527061
- DOI: 10.1021/bi062269m
Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies
Abstract
Base excision repair (BER) is the major pathway employed to excise oxidized DNA lesions. Human Neil1, a versatile glycosylase in the BER pathway, repairs a diverse array of oxidative lesions; however, the most prevalent, 8-oxo-7,8-dihydroguanine (8-oxoG), is only weakly excised. The structural origin of hNeil1's ability to repair a variety of lesions but not 8-oxoG is a model system for connecting enzyme structure and lesion-recognition specificity. To elucidate structural properties determining hNeil1's substrate specificities, we have investigated it in complex with two pairs of representative well-repaired substrates: the R- and S-spiroiminodihydantoin (Sp) stereoisomers, nonplanar further oxidation products of guanine, and the 5R,6S- and 5S,6R-thymine glycol (Tg) stereoisomers, the most prevalent oxidative lesions of thymine. We also investigate the poorly repaired 8-oxoG. We employed molecular modeling and 10 ns molecular dynamics (MD) simulations. The results of our investigations provide structural explanations for the ability of hNeil1 to excise a variety of oxidative lesions: they possess common chemical features, namely, a pyrimidine-like ring and shared hydrogen bond donor-acceptor properties, which allow the lesions to fit well in the binding pocket, which is somewhat flexible. However, the planar 8-oxoG is not as well accommodated in the shallow and comparatively cramped recognition pocket; it has fewer hydrogen bonding interactions with the enzyme and a solvent exposed six-membered ring, consistent with its poor repair susceptibility by this enzyme.
Figures

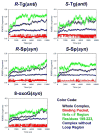

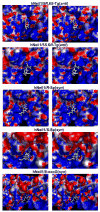


Similar articles
-
Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1.Biochemistry. 2008 Jul 8;47(27):7137-46. doi: 10.1021/bi800160s. Epub 2008 Jun 11. Biochemistry. 2008. PMID: 18543945 Free PMC article.
-
Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2.DNA Repair (Amst). 2005 Jan 2;4(1):41-50. doi: 10.1016/j.dnarep.2004.07.006. DNA Repair (Amst). 2005. PMID: 15533836
-
Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context.J Biol Chem. 2013 Sep 20;288(38):27263-27272. doi: 10.1074/jbc.M113.479055. Epub 2013 Aug 7. J Biol Chem. 2013. PMID: 23926102 Free PMC article.
-
Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases.Free Radic Biol Med. 2017 Jun;107:179-201. doi: 10.1016/j.freeradbiomed.2016.11.042. Epub 2016 Nov 27. Free Radic Biol Med. 2017. PMID: 27903453 Review.
-
Understanding the sequence and structural context effects in oxidative DNA damage repair.DNA Repair (Amst). 2020 Sep;93:102906. doi: 10.1016/j.dnarep.2020.102906. DNA Repair (Amst). 2020. PMID: 33087272 Review.
Cited by
-
Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes containing single diastereomeric spiroiminodihydantoin lesions.Biochemistry. 2013 Feb 26;52(8):1354-63. doi: 10.1021/bi301566v. Epub 2013 Feb 13. Biochemistry. 2013. PMID: 23360616 Free PMC article.
-
The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure.J Biol Chem. 2009 May 1;284(18):11963-70. doi: 10.1074/jbc.M900746200. Epub 2009 Mar 3. J Biol Chem. 2009. PMID: 19258314 Free PMC article.
-
RNA Editing of the Human DNA Glycosylase NEIL1 Alters Its Removal of 5-Hydroxyuracil Lesions in DNA.Biochemistry. 2021 May 18;60(19):1485-1497. doi: 10.1021/acs.biochem.1c00062. Epub 2021 Apr 30. Biochemistry. 2021. PMID: 33929180 Free PMC article.
-
Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5'-GTgG-3' sequence: destabilization of two base pairs at the lesion site.Nucleic Acids Res. 2010 Jan;38(2):428-40. doi: 10.1093/nar/gkp844. Epub 2009 Nov 5. Nucleic Acids Res. 2010. PMID: 19892827 Free PMC article.
-
Influence of substrate complexity on the diastereoselective formation of spiroiminodihydantoin and guanidinohydantoin from chromate oxidation.Chem Res Toxicol. 2010 Feb 15;23(2):379-85. doi: 10.1021/tx900362r. Chem Res Toxicol. 2010. PMID: 20014751 Free PMC article.
References
-
- Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. - PubMed
-
- Scharer OD. Chemistry and biology of DNA repair. Angew Chem, Int Ed. 2003;42:2946–74. - PubMed
-
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. - PubMed
-
- Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–8. - PubMed
-
- David SS. Structural biology: DNA search and rescue. Nature. 2005;434:569–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

