Cisplatin damage overrides the predefined rotational setting of positioned nucleosomes
- PMID: 17432860
- PMCID: PMC2501106
- DOI: 10.1021/ja0706145
Cisplatin damage overrides the predefined rotational setting of positioned nucleosomes
Abstract
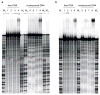
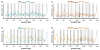
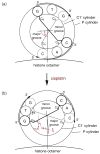
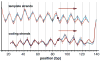
Cisplatin and carboplatin are used successfully to treat various types of cancer. The drugs target the nucleosomes of cancer cells and form intrastrand DNA cross-links that are located in the major groove. We constructed two site-specifically modified nucleosomes containing defined intrastrand cis-{Pt(NH3)2}(2+) 1,3-d(GpTpG) cross-links. Histones from HeLa-S3 cancer cells were transferred onto synthetic DNA duplexes having nucleosome positioning sequences. The structures of these complexes were investigated by hydroxyl radical footprinting. Employing nucleosome positioning sequences allowed us to quantify the structural deviation induced by the cisplatin adduct. Our experiments demonstrate that a platinum cross-link locally overrides the rotational setting predefined in the nucleosome positioning sequence such that the lesion faces toward the histone core. Identical results were obtained for two DNA duplexes in which the sites of platination differed by approximately half a helical turn. Additionally, we determined that cisplatin unwinds nucleosomal DNA globally by approximately 24 degree. The intrastrand cis-{Pt(NH3)2}(2+) 1,3-d(GpTpG) cross-links are located in an area of the nucleosome that contains locally overwound DNA in undamaged reference nucleosomes. Because most nucleosome positions in vivo are defined by the intrinsic DNA sequence, the ability of cisplatin to influence the structure of these positioned nucleosomes may be of physiological relevance.
Figures



References
-
- Wong E, Giandomenico CM. Chem Rev. 1999;99:2451–2466. - PubMed
-
- Jamieson ER, Lippard SJ. Chem Rev. 1999;99:2467–2498. - PubMed
-
- Wang D, Lippard SJ. Nat Rev Drug Discovery. 2005;4:307–319. - PubMed
-
- Blommaert FA, van Dijk-Knijnenburg HCM, Dijt FJ, den Engelse L, Baan RA, Berends F, Fichtinger-Schepman AMJ. Biochemistry. 1995;34:8474–8480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

