Cell size at S phase initiation: an emergent property of the G1/S network
- PMID: 17432928
- PMCID: PMC1851985
- DOI: 10.1371/journal.pcbi.0030064
Cell size at S phase initiation: an emergent property of the G1/S network
Abstract
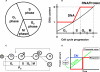
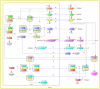
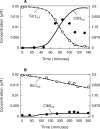
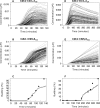
The eukaryotic cell cycle is the repeated sequence of events that enable the division of a cell into two daughter cells. It is divided into four phases: G1, S, G2, and M. Passage through the cell cycle is strictly regulated by a molecular interaction network, which involves the periodic synthesis and destruction of cyclins that bind and activate cyclin-dependent kinases that are present in nonlimiting amounts. Cyclin-dependent kinase inhibitors contribute to cell cycle control. Budding yeast is an established model organism for cell cycle studies, and several mathematical models have been proposed for its cell cycle. An area of major relevance in cell cycle control is the G1 to S transition. In any given growth condition, it is characterized by the requirement of a specific, critical cell size, PS, to enter S phase. The molecular basis of this control is still under discussion. The authors report a mathematical model of the G1 to S network that newly takes into account nucleo/cytoplasmic localization, the role of the cyclin-dependent kinase Sic1 in facilitating nuclear import of its cognate Cdk1-Clb5, Whi5 control, and carbon source regulation of Sic1 and Sic1-containing complexes. The model was implemented by a set of ordinary differential equations that describe the temporal change of the concentration of the involved proteins and protein complexes. The model was tested by simulation in several genetic and nutritional setups and was found to be neatly consistent with experimental data. To estimate PS, the authors developed a hybrid model including a probabilistic component for firing of DNA replication origins. Sensitivity analysis of PS provides a novel relevant conclusion: PS is an emergent property of the G1 to S network that strongly depends on growth rate.
Conflict of interest statement
Figures



References
-
- Mitchison JM. The biology of the cell cycle. Cambridge: Cambridge University Press; 1971. 320
-
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. - PubMed
-
- Grebien F, Dolznig H, Beug H, Mullner EW. Cell size control: New evidence for a general mechanism. Cell Cycle. 2005;4:418–421. - PubMed
-
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

