Molecular basis for a lack of correlation between viral fitness and cell killing capacity
- PMID: 17432933
- PMCID: PMC1851977
- DOI: 10.1371/journal.ppat.0030053
Molecular basis for a lack of correlation between viral fitness and cell killing capacity
Abstract
The relationship between parasite fitness and virulence has been the object of experimental and theoretical studies often with conflicting conclusions. Here, we provide direct experimental evidence that viral fitness and virulence, both measured in the same biological environment provided by host cells in culture, can be two unrelated traits. A biological clone of foot-and-mouth disease virus acquired high fitness and virulence (cell killing capacity) upon large population passages in cell culture. However, subsequent plaque-to-plaque transfers resulted in profound fitness loss, but only a minimal decrease of virulence. While fitness-decreasing mutations have been mapped throughout the genome, virulence determinants-studied here with mutant and chimeric viruses-were multigenic, but concentrated on some genomic regions. Therefore, we propose a model in which viral virulence is more robust to mutation than viral fitness. As a consequence, depending on the passage regime, viral fitness and virulence can follow different evolutionary trajectories. This lack of correlation is relevant to current models of attenuation and virulence in that virus de-adaptation need not entail a decrease of virulence.
Conflict of interest statement
Figures
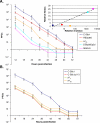
 , C-S8p260p3d, and MARLS to kill 104 BHK-21 cells as a function of the initial number of infectious units (PFU) added. The virulence assay is described in Materials and Methods. Each value represents the mean and standard deviation from triplicate assays. Inset: Relative fitness as a function of relative virulence values of FMDVs. The regression (discontinuous) line defined by C-S8c1, REDpt60, C-S8p260p3d, and MARLS is y = 3.026Ln(x) – 1.1028; R
2 = 0.9721. The regression line including
, C-S8p260p3d, and MARLS to kill 104 BHK-21 cells as a function of the initial number of infectious units (PFU) added. The virulence assay is described in Materials and Methods. Each value represents the mean and standard deviation from triplicate assays. Inset: Relative fitness as a function of relative virulence values of FMDVs. The regression (discontinuous) line defined by C-S8c1, REDpt60, C-S8p260p3d, and MARLS is y = 3.026Ln(x) – 1.1028; R
2 = 0.9721. The regression line including  is y = 3.1458Ln(x) – 3.5111; R
2 = 0.8507 (not drawn). (B) Time needed by C-S8c1, C-S8c1p113,
is y = 3.1458Ln(x) – 3.5111; R
2 = 0.8507 (not drawn). (B) Time needed by C-S8c1, C-S8c1p113,  , and
, and  to kill 104 BHK-21 cells as a function of the initial number of PFU. Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods, and their evolutionary relationship is depicted in Figure 1. Virulence values are given in Tables 1, 2, and S1.
to kill 104 BHK-21 cells as a function of the initial number of PFU. Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods, and their evolutionary relationship is depicted in Figure 1. Virulence values are given in Tables 1, 2, and S1.
 -coding regions are represented in white and blue, respectively. Amino acids in
-coding regions are represented in white and blue, respectively. Amino acids in  that differ from the corresponding ones in C-S8c1 are indicated in red. Replacements found also in the
that differ from the corresponding ones in C-S8c1 are indicated in red. Replacements found also in the  genome are encircled. Numbering of amino acids for each individual protein is as in [11].
genome are encircled. Numbering of amino acids for each individual protein is as in [11].
 regulatory regions are represented as black and blue lines, respectively; the corresponding coding regions are represented as white and blue boxes, respectively. pMT28 refers to the plasmid that encodes infectious RNA transcripts with the C-S8c1 nucleotide sequence [22]. Chimeric viruses are colored blue and named with the first and the last nucleotide that correspond to the
regulatory regions are represented as black and blue lines, respectively; the corresponding coding regions are represented as white and blue boxes, respectively. pMT28 refers to the plasmid that encodes infectious RNA transcripts with the C-S8c1 nucleotide sequence [22]. Chimeric viruses are colored blue and named with the first and the last nucleotide that correspond to the  genomic region. Chimera
genomic region. Chimera  /2C-3A(pMT28) includes 2C, 3A, and parts of 2B and 3B of pMT28 in the background of residues 436 to 7427 of
/2C-3A(pMT28) includes 2C, 3A, and parts of 2B and 3B of pMT28 in the background of residues 436 to 7427 of  . Amino acid substitutions in protein 2C are indicated in red. The residue numbering of the FMDV genome is as in [11]. Procedures for the construction of chimeric viruses and 2C mutants are described in Materials and Methods.
. Amino acid substitutions in protein 2C are indicated in red. The residue numbering of the FMDV genome is as in [11]. Procedures for the construction of chimeric viruses and 2C mutants are described in Materials and Methods.
 , pMT28/
, pMT28/ (436-2046), pMT28/
(436-2046), pMT28/ (2046–3760), and pMT28/
(2046–3760), and pMT28/ (436-3760). (B) C-S8c1(pMT28),
(436-3760). (B) C-S8c1(pMT28),  , pMT28/
, pMT28/ (3760–5839), pMT28/
(3760–5839), pMT28/ (5839–7427), and pMT28/
(5839–7427), and pMT28/ (3760–7427). (C) C-S8c1(pMT28),
(3760–7427). (C) C-S8c1(pMT28),  , pMT28/
, pMT28/ (2046–7427), and pMT28/
(2046–7427), and pMT28/ (436-7427). (D) C-S8c1(pMT28),
(436-7427). (D) C-S8c1(pMT28),  , pMT28/
, pMT28/ (436-7427), and
(436-7427), and  /2C-3A(pMT28). Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods; C-S8c1(pMT28) is C-S8c1 expressed as an infectious transcript and rescued by cell transfection. Virulence values are given in Tables 2 and S1.
/2C-3A(pMT28). Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods; C-S8c1(pMT28) is C-S8c1 expressed as an infectious transcript and rescued by cell transfection. Virulence values are given in Tables 2 and S1.
 , pMT28 (SN), pMT28 (TA), and pMT28 (QH). (B) C-S8c1(pMT28),
, pMT28 (SN), pMT28 (TA), and pMT28 (QH). (B) C-S8c1(pMT28),  , and pMT28 (SN, TA, QH). Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods. Virulence values are given in Tables 2 and S1.
, and pMT28 (SN, TA, QH). Each value represents the mean and standard deviation from triplicate assays. The viruses analyzed are described in Materials and Methods. Virulence values are given in Tables 2 and S1.References
-
- Domingo E, Biebricher C, Eigen M, Holland JJ. Quasispecies and RNA virus evolution: Principles and consequences. Austin (Texas): Landes Bioscience; 2001. 221
-
- Biebricher CK, Eigen M. What is a quasispecies? Curr Top Microbiol Immunol. 2006;299:1–31. - PubMed
-
- Eigen M, Biebricher CK. Sequence space and quasispecies distribution. In: Domingo E, Ahlquist P, Holland JJ, editors. RNA genetics. Boca Raton (Florida): CRC Press; 1988. pp. 211–245.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

