HHV-8 encoded LANA-1 alters the higher organization of the cell nucleus
- PMID: 17433107
- PMCID: PMC1857702
- DOI: 10.1186/1476-4598-6-28
HHV-8 encoded LANA-1 alters the higher organization of the cell nucleus
Abstract
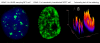
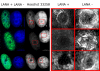
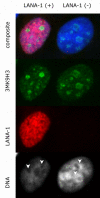
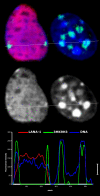
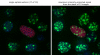
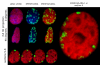
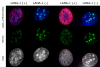
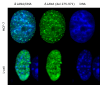
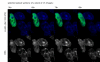
The latency-associated nuclear antigen (LANA-1) of Human Herpes Virus 8 (HHV-8), alternatively called Kaposi Sarcoma Herpes Virus (KSHV) is constitutively expressed in all HHV-8 infected cells. LANA-1 accumulates in well-defined foci that co-localize with the viral episomes. We have previously shown that these foci are tightly associated with the borders of heterochromatin 1. We have also shown that exogenously expressed LANA-1 causes an extensive re-organization of Hoechst 33248 DNA staining patterns of the nuclei in non-HHV-8 infected cells 2. Here we show that this effect includes the release of the bulk of DNA from heterochromatic areas, in both human and mouse cells, without affecting the overall levels of heterochromatin associated histone H3 lysine 9 tri-methylation (3MK9H3). The release of DNA from the heterochromatic chromocenters in LANA-1 transfected mouse cells co-incides with the dispersion of the chromocenter associated methylcytosin binding protein 2 (MECP2). The localization of 3MK9H3 to the remnants of the chromocenters remains unaltered. Moreover, exogeneously expressed LANA-1 leads to the relocation of the chromocenters to the nuclear periphery, indicating extensive changes in the positioning of the chromosomal domains in the LANA-1 harboring interphase nucleus. Using a series of deletion mutants we have shown that the chromatin rearranging effects of LANA-1 require the presence of a short (57 amino acid) region that is located immediately upstream of the internal acidic repeats. This sequence lies within the previously mapped binding site to histone methyltransferase SUV39H1. We suggest that the highly concentrated LANA-1, anchored to the host genome in the nuclear foci of latently infected cells and replicated through each cell generation, may function as "epigenetic modifier". The induction of histone modification in adjacent host genes may lead to altered gene expression, thereby contributing to the viral oncogenesis.
Figures



Similar articles
-
The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains.J Virol. 2010 Mar;84(5):2318-30. doi: 10.1128/JVI.01097-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032179 Free PMC article.
-
Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen.J Virol. 2004 Jul;78(14):7299-310. doi: 10.1128/JVI.78.14.7299-7310.2004. J Virol. 2004. PMID: 15220403 Free PMC article.
-
The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells.Virology. 1999 Nov 25;264(2):254-64. doi: 10.1006/viro.1999.9999. Virology. 1999. PMID: 10562490
-
In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus.Mol Immunol. 2007 Feb;44(6):1352-60. doi: 10.1016/j.molimm.2006.05.012. Epub 2006 Jul 7. Mol Immunol. 2007. PMID: 16828498
-
Accumulation of LANA at nuclear matrix fraction is important for Kaposi's sarcoma-associated herpesvirus replication in latency.Virus Res. 2009 Jan;139(1):74-84. doi: 10.1016/j.virusres.2008.10.011. Epub 2008 Dec 9. Virus Res. 2009. PMID: 19027806
Cited by
-
LANA-1, Bcl-2, Mcl-1 and HIF-1alpha protein expression in HIV-associated Kaposi sarcoma.Virchows Arch. 2009 Aug;455(2):159-70. doi: 10.1007/s00428-009-0791-1. Epub 2009 May 30. Virchows Arch. 2009. PMID: 19484260
-
Extended Field Laser Confocal Microscopy (EFLCM): combining automated Gigapixel image capture with in silico virtual microscopy.BMC Med Imaging. 2008 Jul 16;8:13. doi: 10.1186/1471-2342-8-13. BMC Med Imaging. 2008. PMID: 18627634 Free PMC article.
-
Epstein-Barr virus-mediated transformation of B cells induces global chromatin changes independent to the acquisition of proliferation.Nucleic Acids Res. 2014 Jan;42(1):249-63. doi: 10.1093/nar/gkt886. Epub 2013 Oct 3. Nucleic Acids Res. 2014. PMID: 24097438 Free PMC article.
-
The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains.J Virol. 2010 Mar;84(5):2318-30. doi: 10.1128/JVI.01097-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032179 Free PMC article.
-
KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor.Blood. 2008 May 1;111(9):4731-40. doi: 10.1182/blood-2007-09-110544. Epub 2008 Jan 16. Blood. 2008. PMID: 18199825 Free PMC article.
References
-
- Szekely L, Kiss C, Mattsson K, Kashuba E, Pokrovskaja K, Juhasz A, Holmvall P, Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80:2889–2900. - PubMed
-
- Mattsson K, Kiss C, Platt GM, Simpson GR, Kashuba E, Klein G, Schulz TF, Szekely L. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J Gen Virol. 2002;83:179–188. - PubMed
-
- Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, Hardwick JM. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

