The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains
- PMID: 17434555
- PMCID: PMC1950718
- DOI: 10.1016/j.virol.2007.03.019
The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains
Abstract
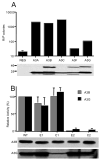
The mammalian APOBEC3 proteins are cytidine deaminases that function as inhibitors of retrovirus replication and retrotransposon mobility. An issue that has remained controversial is whether the editing of deoxycytidine residues to deoxyuridine is necessary and sufficient for this inhibition or whether APOBEC3 proteins also exert a second, distinct inhibitory mechanism. Here, we present an analysis of the ability of mutants of APOBEC3G and APOBEC3B, both of which contain two consensus cytidine deaminase active sites, to inhibit the replication of human immunodeficiency virus. Our data confirm that APOBEC3G only contains a single, carboxy-terminal active site but, surprisingly, reveal that both cytidine deaminase consensus sequences in APOBEC3B are enzymatically active. Enzymatically inactive mutant forms of APOBEC3G and APOBEC3B were found to retain the ability to inhibit the infectivity of HIV-1 virions produced in their presence by approximately 4-fold and approximately 8-fold, respectively. While this inhibition was significantly less than the level seen with wild-type forms of A3G or A3B, these data, nevertheless argue that the inhibition of HIV-1 by APOBEC3 proteins is at least partly independent of DNA editing.
Figures


Similar articles
-
Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B.J Virol. 2006 Apr;80(7):3679-83. doi: 10.1128/JVI.80.7.3679-3683.2006. J Virol. 2006. PMID: 16537639 Free PMC article.
-
Functional domain organization of human APOBEC3G.Virology. 2008 Sep 15;379(1):118-24. doi: 10.1016/j.virol.2008.06.013. Epub 2008 Jul 18. Virology. 2008. PMID: 18639915 Free PMC article.
-
Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities.Nucleic Acids Res. 2005 Apr 4;33(6):1913-23. doi: 10.1093/nar/gki343. Print 2005. Nucleic Acids Res. 2005. PMID: 15809227 Free PMC article.
-
APOBEC deaminases as cellular antiviral factors: a novel natural host defense mechanism.Med Sci Monit. 2006 May;12(5):RA92-8. Med Sci Monit. 2006. PMID: 16641889 Review.
-
Multifaceted antiviral actions of APOBEC3 cytidine deaminases.Trends Immunol. 2006 Jun;27(6):291-7. doi: 10.1016/j.it.2006.04.003. Epub 2006 May 4. Trends Immunol. 2006. PMID: 16678488 Review.
Cited by
-
Protein kinase A inhibits tumor mutator APOBEC3B through phosphorylation.Sci Rep. 2019 Jun 5;9(1):8307. doi: 10.1038/s41598-019-44407-9. Sci Rep. 2019. PMID: 31165764 Free PMC article.
-
miR-128 Restriction of LINE-1 (L1) Retrotransposition Is Dependent on Targeting hnRNPA1 mRNA.Int J Mol Sci. 2019 Apr 21;20(8):1955. doi: 10.3390/ijms20081955. Int J Mol Sci. 2019. PMID: 31010097 Free PMC article.
-
The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B.J Virol. 2011 Sep;85(17):8538-47. doi: 10.1128/JVI.02645-10. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715505 Free PMC article.
-
D316 is critical for the enzymatic activity and HIV-1 restriction potential of human and rhesus APOBEC3B.Virology. 2013 Jun 20;441(1):31-9. doi: 10.1016/j.virol.2013.03.003. Epub 2013 Mar 29. Virology. 2013. PMID: 23542011 Free PMC article.
-
Nuclear Magnetic Resonance Structure of the APOBEC3B Catalytic Domain: Structural Basis for Substrate Binding and DNA Deaminase Activity.Biochemistry. 2016 May 31;55(21):2944-59. doi: 10.1021/acs.biochem.6b00382. Epub 2016 May 19. Biochemistry. 2016. PMID: 27163633 Free PMC article.
References
-
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. - PubMed
-
- Beale RCL, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. - PubMed
-
- Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr Cytidine deaminase. The 2·3 Å crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994;235:635–656. - PubMed
-
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current Biol. 2004;14:1392–1396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

