Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii
- PMID: 17435007
- PMCID: PMC1951523
- DOI: 10.1128/EC.00207-06
Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii
Abstract
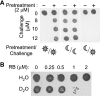
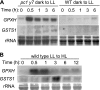
In an aerobic environment, responding to oxidative cues is critical for physiological adaptation (acclimation) to changing environmental conditions. The unicellular alga Chlamydomonas reinhardtii was tested for the ability to acclimate to specific forms of oxidative stress. Acclimation was defined as the ability of a sublethal pretreatment with a reactive oxygen species to activate defense responses that subsequently enhance survival of that stress. C. reinhardtii exhibited a strong acclimation response to rose bengal, a photosensitizing dye that produces singlet oxygen. This acclimation was dependent upon photosensitization and occurred only when pretreatment was administered in the light. Shifting cells from low light to high light also enhanced resistance to singlet oxygen, suggesting an overlap in high-light and singlet oxygen response pathways. Microarray analysis of RNA levels indicated that a relatively small number of genes respond to sublethal levels of singlet oxygen. Constitutive overexpression of either of two such genes, a glutathione peroxidase gene and a glutathione S-transferase gene, was sufficient to enhance singlet oxygen resistance. Escherichia coli and Saccharomyces cerevisiae exhibit well-defined responses to reactive oxygen but did not acclimate to singlet oxygen, possibly reflecting the relative importance of singlet oxygen stress for photosynthetic organisms.
Figures








References
-
- Agianian, B., P. A. Tucker, A. Schouten, K. Leonard, B. Bullard, and P. Gros. 2003. Structure of a Drosophila sigma class glutathione S-transferase reveals a novel active site topography suited for lipid peroxidation products. J. Mol. Biol. 326:151-165. - PubMed
-
- Alic, N., T. Felder, M. D. Temple, C. Gloeckner, V. J. Higgins, P. Briza, and I. W. Dawes. 2004. Genome-wide transcriptional responses to a lipid hydroperoxide: adaptation occurs without induction of oxidant defenses. Free Radic. Biol. Med. 37:23-35. - PubMed
-
- Apel, K., and H. Hirt. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373-399. - PubMed
-
- Avery, A. M., and S. V. Avery. 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276:33730-33735. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

