Bayesian shrinkage analysis of quantitative trait Loci for dynamic traits
- PMID: 17435239
- PMCID: PMC1894582
- DOI: 10.1534/genetics.106.064279
Bayesian shrinkage analysis of quantitative trait Loci for dynamic traits
Abstract
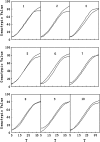
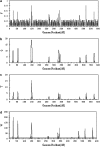
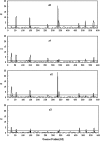
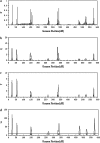
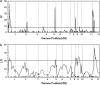
Many quantitative traits are measured repeatedly during the life of an organism. Such traits are called dynamic traits. The pattern of the changes of a dynamic trait is called the growth trajectory. Studying the growth trajectory may enhance our understanding of the genetic architecture of the growth trajectory. Recently, we developed an interval-mapping procedure to map QTL for dynamic traits under the maximum-likelihood framework. We fit the growth trajectory by Legendre polynomials. The method intended to map one QTL at a time and the entire QTL analysis involved scanning the entire genome by fitting multiple single-QTL models. In this study, we propose a Bayesian shrinkage analysis for estimating and mapping multiple QTL in a single model. The method is a combination between the shrinkage mapping for individual quantitative traits and the Legendre polynomial analysis for dynamic traits. The multiple-QTL model is implemented in two ways: (1) a fixed-interval approach where a QTL is placed in each marker interval and (2) a moving-interval approach where the position of a QTL can be searched in a range that covers many marker intervals. Simulation study shows that the Bayesian shrinkage method generates much better signals for QTL than the interval-mapping approach. We propose several alternative methods to present the results of the Bayesian shrinkage analysis. In particular, we found that the Wald test-statistic profile can serve as a mechanism to test the significance of a putative QTL.
Figures





Similar articles
-
Bayesian B-spline mapping for dynamic quantitative traits.Genet Res (Camb). 2012 Apr;94(2):85-95. doi: 10.1017/S0016672312000249. Genet Res (Camb). 2012. PMID: 22624568
-
Bayesian analysis for genetic architecture of dynamic traits.Heredity (Edinb). 2011 Jan;106(1):124-33. doi: 10.1038/hdy.2010.20. Epub 2010 Mar 24. Heredity (Edinb). 2011. PMID: 20332806 Free PMC article.
-
[Maximum likelihood analysis for mapping dynamic trait QTL in outbred population. I . Methodology].Yi Chuan Xue Bao. 2004 Oct;31(10):1116-22. Yi Chuan Xue Bao. 2004. PMID: 15552047 Chinese.
-
Advances in Bayesian multiple quantitative trait loci mapping in experimental crosses.Heredity (Edinb). 2008 Mar;100(3):240-52. doi: 10.1038/sj.hdy.6801074. Epub 2007 Nov 7. Heredity (Edinb). 2008. PMID: 17987056 Free PMC article. Review.
-
Current progress on statistical methods for mapping quantitative trait loci from inbred line crosses.J Biopharm Stat. 2010 Mar;20(2):454-81. doi: 10.1080/10543400903572845. J Biopharm Stat. 2010. PMID: 20309768 Review.
Cited by
-
A Gaussian process model and Bayesian variable selection for mapping function-valued quantitative traits with incomplete phenotypic data.Bioinformatics. 2019 Oct 1;35(19):3684-3692. doi: 10.1093/bioinformatics/btz164. Bioinformatics. 2019. PMID: 30850830 Free PMC article.
-
Robust Bayesian mapping of quantitative trait loci using Student-t distribution for residual.Theor Appl Genet. 2009 Feb;118(3):609-17. doi: 10.1007/s00122-008-0924-y. Epub 2008 Nov 20. Theor Appl Genet. 2009. PMID: 19020853
-
A new mapping method for quantitative trait loci of silkworm.BMC Genet. 2011 Jan 28;12:19. doi: 10.1186/1471-2156-12-19. BMC Genet. 2011. PMID: 21276233 Free PMC article.
-
Methodologies for segregation analysis and QTL mapping in plants.Genetica. 2009 Jun;136(2):311-8. doi: 10.1007/s10709-008-9313-3. Epub 2008 Aug 23. Genetica. 2009. PMID: 18726162 Review.
-
BAYESIAN GROUP LASSO FOR NONPARAMETRIC VARYING-COEFFICIENT MODELS WITH APPLICATION TO FUNCTIONAL GENOME-WIDE ASSOCIATION STUDIES.Ann Appl Stat. 2015 Jun;9(2):640-664. doi: 10.1214/15-AOAS808. Ann Appl Stat. 2015. PMID: 26478762 Free PMC article.
References
-
- Bink, M. C. A. M., P. Uimari, M. J. Sillanpää, L. L. G. Janss and R. C. Jansen, 2002. Multiple QTL mapping in related plant populations via a pedigree-analysis approach. Theor. Appl. Genet. 104 751–762. - PubMed
-
- Corander, J., and M. J. Sillanpää, 2002. A unified approach to joint modeling of multiple quantitative and qualitative traits in gene mapping. J. Theor. Biol. 218 435–446. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

