Involvement of the mitochondrial protein translocator component tim50 in growth, cell proliferation and the modulation of respiration in Drosophila
- PMID: 17435247
- PMCID: PMC1894619
- DOI: 10.1534/genetics.107.072074
Involvement of the mitochondrial protein translocator component tim50 in growth, cell proliferation and the modulation of respiration in Drosophila
Abstract
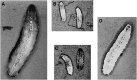
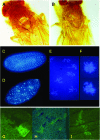
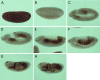
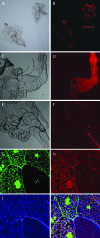
Allelic mutants exhibiting growth defects in Drosophila were isolated. Molecular cloning identified the responsible gene as a budding yeast Tim50 ortholog, and thus it was named tiny tim 50 (ttm50). The weak allele (ttm50(Gp99)) produced small flies due to reduced cell size and number, and growth terminated at the larval stage in the strong alleles (ttm50(IE1) and ttm50(IE2)). Twin-spot analysis showed fewer cells in ttm50(Gp99) clones, whereas ttm50(IE1) clones did not proliferate, suggesting that the gene has an essential cellular function. Tim50 is known to maintain mitochondrial membrane potential (MMP) while facilitating inner-membrane protein transport. We found that tagged Ttm50 also localized to mitochondria and that mitochondrial morphology and MMP were affected in mutants, indicating that mitochondrial dysfunction causes the developmental phenotype. Conversely, ttm50 overexpression increased MMP and apoptosis. Co-expression of p35 suppressed this apoptosis, resulting in cell overproliferation. Interestingly, ttm50 transcription was tissue specific, corresponding to elevated MMP in the larval midgut, which was decreased in the mutant. The correlation of ttm50 expression levels with differences in MMP match its proposed role in mitochondrial permeability barrier maintenance. Thus a mitochondrial protein translocase component can play active roles in regulating metabolic levels, possibly for modulation of physiological function or growth in development.
Figures


Similar articles
-
Transcriptional regulation of the Drosophila ANT gene by the DRE/DREF system.Genes Cells. 2007 May;12(5):569-79. doi: 10.1111/j.1365-2443.2007.01075.x. Genes Cells. 2007. PMID: 17535248
-
Ttm50 facilitates calpain activation by anchoring it to calcium stores and increasing its sensitivity to calcium.Cell Res. 2021 Apr;31(4):433-449. doi: 10.1038/s41422-020-0388-4. Epub 2020 Aug 26. Cell Res. 2021. PMID: 32848200 Free PMC article.
-
humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila.Curr Biol. 2005 Apr 26;15(8):755-9. doi: 10.1016/j.cub.2005.02.063. Curr Biol. 2005. PMID: 15854909
-
The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling.J Biol. 2003;2(3):20. doi: 10.1186/1475-4924-2-20. Epub 2003 Aug 7. J Biol. 2003. PMID: 12908874 Free PMC article.
-
Increased synaptic microtubules and altered synapse development in Drosophila sec8 mutants.BMC Biol. 2005 Dec 13;3:27. doi: 10.1186/1741-7007-3-27. BMC Biol. 2005. PMID: 16351720 Free PMC article.
Cited by
-
A mitochondrial redox switch licenses the onset of morphogenesis in animals.bioRxiv [Preprint]. 2024 Oct 29:2024.10.28.620733. doi: 10.1101/2024.10.28.620733. bioRxiv. 2024. PMID: 39553983 Free PMC article. Preprint.
-
Loss of TIM50 suppresses proliferation and induces apoptosis in breast cancer.Tumour Biol. 2016 Jan;37(1):1279-87. doi: 10.1007/s13277-015-3878-0. Epub 2015 Aug 20. Tumour Biol. 2016. PMID: 26289846
-
Invadolysin, a conserved lipid-droplet-associated metalloproteinase, is required for mitochondrial function in Drosophila.J Cell Sci. 2013 Oct 15;126(Pt 20):4769-81. doi: 10.1242/jcs.133306. Epub 2013 Aug 13. J Cell Sci. 2013. PMID: 23943867 Free PMC article.
-
Mutations in the Drosophila mitochondrial tRNA amidotransferase, bene/gatA, cause growth defects in mitotic and endoreplicating tissues.Genetics. 2008 Feb;178(2):979-87. doi: 10.1534/genetics.107.084376. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245325 Free PMC article.
-
The Cross Talk between TbTim50 and PIP39, Two Aspartate-Based Protein Phosphatases, Maintains Cellular Homeostasis in Trypanosoma brucei.mSphere. 2019 Aug 7;4(4):e00353-19. doi: 10.1128/mSphere.00353-19. mSphere. 2019. PMID: 31391278 Free PMC article.
References
-
- Böhni, R., J. Riesgo-Escovar, S. Oldham, W. Brogiolo, H. Stocker et al., 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97 865–875. - PubMed
-
- Brand, A. H., and N. Perrimon, 1993. Targeted expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. - PubMed
-
- Brown, N. H., and F. C. Kafatos, 1988. Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203 425–437. - PubMed
-
- Chen, C., J. Joseph and R. S. Garofalo, 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137 846–856. - PubMed
-
- Curran, S, P., E. P. Leverich, C. M. Koehler and P. L. Larsen, 2004. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J. Biol. Chem. 279 54655–54662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

