Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities
- PMID: 17436248
- PMCID: PMC1852738
- DOI: 10.1086/513607
Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities
Abstract
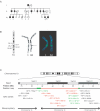
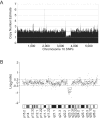
Low-copy repeats (LCRs) are genomic features that affect chromosome stability and can produce disease-associated rearrangements. We describe members of three families with deletions in 10q22.3-q23.31, a region harboring a complex set of LCRs, and demonstrate that rearrangements in this region are associated with behavioral and neurodevelopmental abnormalities, including cognitive impairment, autism, hyperactivity, and possibly psychiatric disease. Fine mapping of the deletions in members of all three families by use of a custom 10q oligonucleotide array-based comparative genomic hybridization (NimbleGen) and polymerase chain reaction-based methods demonstrated a different deletion in each family. In one proband, the deletion breakpoints are associated with DNA fragments containing noncontiguous sequences of chromosome 10, whereas, in the other two families, the breakpoints are within paralogous LCRs, removing approximately 7.2 Mb and 32 genes. Our data provide evidence that the 10q22-q23 genomic region harbors one or more genes important for cognitive and behavioral development and that recurrent deletions affecting this interval define a novel genomic disorder.
Figures



References
Web Resources
-
- CHORI BACPAC Resources, http://bacpac.chori.org/genomicRearrays.php
-
- Database of Genomic Variants, http://projects.tcag.ca/variation/
-
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/index.html (for GHITM [accession number 17281])
-
- Human Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway (for March 2006 assembly)
-
- Human Genome Segmental Duplication Database, http://projects.tcag.ca/humandup/
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

