Structural basis for selective inhibition of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB
- PMID: 17437721
- PMCID: PMC2775457
- DOI: 10.1016/j.str.2007.03.003
Structural basis for selective inhibition of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB
Abstract
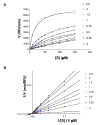
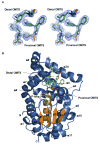
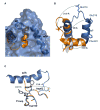
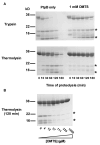
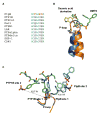
Tyrosine kinases and phosphatases establish the crucial balance of tyrosine phosphorylation in cellular signaling, but creating specific inhibitors of protein Tyr phosphatases (PTPs) remains a challenge. Here, we report the development of a potent, selective inhibitor of Mycobacterium tuberculosis PtpB, a bacterial PTP that is secreted into host cells where it disrupts unidentified signaling pathways. The inhibitor, (oxalylamino-methylene)-thiophene sulfonamide (OMTS), showed an IC(50) of 440 +/- 50 nM and >60-fold specificity for PtpB over six human PTPs. The 2 A resolution crystal structure of PtpB in complex with OMTS revealed a large rearrangement of the enzyme, with some residues shifting >27 A relative to the PtpB:PO(4) complex. Extensive contacts with the catalytic loop provide a potential basis for inhibitor selectivity. Two OMTS molecules bound adjacent to each other, raising the possibility of a second substrate phosphotyrosine binding site in PtpB. The PtpB:OMTS structure provides an unanticipated framework to guide inhibitor improvement.
Figures



References
-
- Andersen HS, Iversen LF, Jeppesen CB, Branner S, Norris K, Rasmussen HB, Moller KB, Moller NP. 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases. J Biol Chem. 2000;275:7101–7108. - PubMed
-
- Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. - PubMed
-
- Bialy L, Waldmann H. Inhibitors of protein tyrosine phosphatases: next-generation drugs? Angew Chem Int Ed Engl. 2005;44:3814–3839. - PubMed
-
- DeVinney I, Steele-Mortimer I, Finlay BB. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 2000;8:29–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

