SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster
- PMID: 17438002
- PMCID: PMC1847716
- DOI: 10.1101/gad.1506607
SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster
Abstract
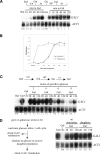
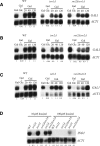
Post-translational modification of nucleosomal histones has been suggested to contribute to epigenetic transcriptional memory. We describe a case of transcriptional memory in yeast where the rate of transcriptional induction of GAL1 is regulated by the prior expression state. This epigenetic state is inherited by daughter cells, but does not require the histone acetyltransferase, Gcn5p, the histone ubiquitinylating enzyme, Rad6p, or the histone methylases, Dot1p, Set1p, or Set2p. In contrast, we show that the ATP-dependent chromatin remodeling enzyme, SWI/SNF, is essential for transcriptional memory at GAL1. Genetic studies indicate that SWI/SNF controls transcriptional memory by antagonizing ISWI-like chromatin remodeling enzymes.
Figures



References
-
- Acar M., Becskei A., van Oudenaarden A., Becskei A., van Oudenaarden A., van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. - PubMed
-
- Becker P.B., Horz W., Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. - PubMed
-
- Briggs S.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Davie J.K., Dent S.Y., Winston F., Allis C.D., Dent S.Y., Winston F., Allis C.D., Winston F., Allis C.D., Allis C.D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
