Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications
- PMID: 17438673
- PMCID: PMC1905836
Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications
Abstract
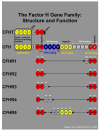
Background: Variants in the complement factor H gene (CFH) are associated with age-related macular degeneration (AMD). CFH and five CFH-related genes (CFHR1-5) lie within the regulators of complement activation (RCA) locus on chromosome 1q32. Aims and Methods. In this study, the structural and evolutionary relationships between these genes and AMD was refined using a combined genetic, molecular and immunohistochemical approach.
Results: We identify and characterize a large, common deletion that encompasses both the CFHR1 and CFHR3 genes. CFHR1, an abundant serum protein, is absent in subjects homozygous for the deletion. Genotyping analyses of AMD cases and controls from two cohorts demonstrates that deletion homozygotes comprise 1.1% of cases and 5.7% of the controls (chi-square=32.8; P= 1.6 E-09). CFHR1 and CFHR3 transcripts are abundant in liver, but undetectable in the ocular retinal pigmented epithelium/choroid complex. AMD-associated CFH/CFHR1/CFHR3 haplotypes are widespread in human populations.
Conclusion: The absence of CFHR1 and/or CFHR3 may account for the protective effects conferred by some CFH haplotypes. Moreover, the high frequencies of the 402H allele and the delCFHR1/CFHR3 alleles in African populations suggest an ancient origin for these alleles. The considerable diversity accumulated at this locus may be due to selection, which is consistent with an important role for the CFHR genes in innate immunity.
Figures


References
-
- Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004 Mar;137(3):486–95. - PubMed
-
- van Leeuwen R, Klaver CC, Vingerling JR, Hofman A, de Jong PT. Epidemiology of age-related maculopathy: a review. Eur J Epidemiol. 2003;18(9):845–54. - PubMed
-
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001 Nov;20(6):705–32. - PubMed
-
- Anderson D, Mullins R, Hageman G, LV J. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
