Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain
- PMID: 17439640
- PMCID: PMC1863423
- DOI: 10.1186/1471-2148-7-62
Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain
Abstract
Background: In plants the hormone cytokinin is perceived by members of a small cytokinin receptor family, which are hybrid sensor histidine kinases. While the immediate downstream signaling pathway is well characterized, the domain of the receptor responsible for ligand binding and which residues are involved in this process has not been determined experimentally.
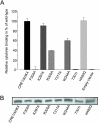
Results: Using a live cell hormone-binding assay, we show that cytokinin is bound by a receptor domain predicted to be extracellular, the so called CHASE (cyclases, histidine kinase associated sensory extracellular) domain. The CHASE domain occurs not only in plant cytokinin receptors but also in numerous orphan receptors in lower eukaryotes and bacteria. Taking advantage of this fact, we used an evolutionary proteomics approach to identify amino acids important for cytokinin binding by looking for residues conserved in cytokinin receptors, but not in other receptors. By comparing differences in evolutionary rates, we predicted five amino acids within the plant CHASE domains to be crucial for cytokinin binding. Mutagenesis of the predicted sites and subsequent binding assays confirmed the relevance of four of the selected amino acids, showing the biological significance of site-specific evolutionary rate differences.
Conclusion: This work demonstrates the use of a bioinformatic analysis to mine the huge set of genomic data from different taxa in order to generate a testable hypothesis. We verified the hypothesis experimentally and identified four amino acids which are to a different degree required for ligand-binding of a plant hormone receptor.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

