The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio
- PMID: 17439641
- PMCID: PMC1867822
- DOI: 10.1186/1471-2148-7-63
The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio
Abstract
Background: The metzincins are a large gene superfamily of proteases characterized by the presence of a zinc protease domain, and include the ADAM, ADAMTS, BMP1/TLL, meprin and MMP genes. Metzincins are involved in the proteolysis of a wide variety of proteins, including those of the extracellular matrix. The metzincin gene superfamily comprises eighty proteins in the human genome and ninety-three in the mouse. When and how the level of complexity apparent in the vertebrate metzincin gene superfamily arose has not been determined in detail. Here we present a comprehensive analysis of vertebrate metzincins using genes from both Ciona intestinalis and Danio rerio to provide new insights into the complex evolution of this gene superfamily.
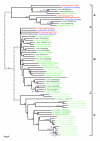
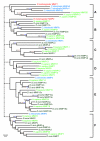
Results: We have identified 19 metzincin genes in the ciona genome and 83 in the zebrafish genome. Phylogenetic analyses reveal that the expansion of the metzincin gene superfamily in vertebrates has occurred predominantly by the simple duplication of pre-existing genes rather than by the appearance and subsequent expansion of new metzincin subtypes (the only example of which is the meprin gene family). Despite the number of zebrafish metzincin genes being relatively similar to that of tetrapods (e.g. man and mouse), the pattern of gene retention and loss within these lineages is markedly different. In addition, we have studied the evolution of the related TIMP gene family and identify a single ciona and four zebrafish TIMP genes.
Conclusion: The complexity seen in the vertebrate metzincin gene families was mainly acquired during vertebrate evolution. The metzincin gene repertoire in protostomes and invertebrate deuterostomes has remained relatively stable. The expanded metzincin gene repertoire of extant tetrapods, such as man, has resulted largely from duplication events associated with early vertebrate evolution, prior to the sarcopterygian-actinopterygian split. The teleost repertoire of metzincin genes in part parallels that of tetrapods but has been significantly modified, perhaps as a consequence of a teleost-specific duplication event.
Figures


References
-
- Bode W., Gomis-Ruth, F-X., Stockler, W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the metzincins. FEBS Letters. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-I. - DOI - PubMed
-
- Black RA., Rauch, C.T., Kozlosky, C.J., Peschon, J.J., Slack, J.L., Wolfson, M.F., Castner, B.J., Stocking, K.L., Reddy, P., Srinivasan, S., Nelson, N., Boiani, N., Schooley, K.A., Gerhart, M., Davis, R., Fitzner, J.N., Johnson, R.S., Paxton, R.J., March, C.J., Cerretti, D.P. A metalloprotease disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

