Pathogenic chaperone-like RNA induces congophilic aggregates and facilitates neurodegeneration in Drosophila
- PMID: 17441503
- PMCID: PMC1874921
- DOI: 10.1379/csc-222r.1
Pathogenic chaperone-like RNA induces congophilic aggregates and facilitates neurodegeneration in Drosophila
Abstract
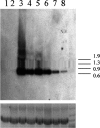
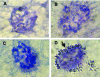
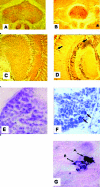
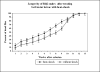
Protein aggregation is a hallmark of many neurodegenerative diseases. RNA chaperones have been suggested to play a role in protein misfolding and aggregation. Noncoding, highly structured RNA recently has been demonstrated to facilitate transformation of recombinant and cellular prion protein into proteinase K-resistant, congophilic, insoluble aggregates and to generate cytotoxic oligomers in vitro. Transgenic Drosophila melanogaster strains were developed to express highly structured RNA under control of a heat shock promoter. Expression of a specific construct strongly perturbed fly behavior, caused significant decline in learning and memory retention of adult males, and was coincident with the formation of intracellular congophilic aggregates in the brain and other tissues of adult and larval stages. Additionally, neuronal cell pathology of adult flies was similar to that observed in human Parkinson's and Alzheimer's disease. This novel model demonstrates that expression of a specific highly structured RNA alone is sufficient to trigger neurodegeneration, possibly through chaperone-like facilitation of protein misfolding and aggregation.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
