GroEL/S substrate specificity based on substrate unfolding propensity
- PMID: 17441504
- PMCID: PMC1852890
- DOI: 10.1379/csc-219r.1
GroEL/S substrate specificity based on substrate unfolding propensity
Abstract
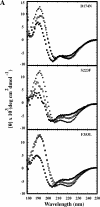
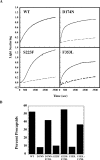
Phage P22 wild-type (WT) coat protein does not require GroEL/S to fold but temperature-sensitive-folding (tsf) coat proteins need the chaperone complex for correct folding. WT coat protein and all variants absolutely require P22 scaffolding protein, an assembly chaperone, to assemble into precursor structures termed procapsids. Previously, we showed that a global suppressor (su) substitution, T1661, which rescues several tsf coat protein variants, functioned by inducing GroEL/S. This led to an increased formation of tsf:T1661 coat protein:GroEL complexes compared with the tsf parents. The increased concentration of complexes resulted in more assembly-competent coat proteins because of a shift in the chaperone-driven kinetic partitioning between aggregation-prone intermediates toward correct folding and assembly. We have now investigated the folding and assembly of coat protein variants that carry a different global su substitution, F170L. By monitoring levels of phage production in the presence of a dysfunctional GroEL we found that tsf:F170L proteins demonstrate a less stringent requirement for GroEL. Tsf:F170L proteins also did not cause induction of the chaperones. Circular dichroism and tryptophan fluorescence indicate that the native state of the tsf: F170L coat proteins is restored to WT-like values. In addition, native acrylamide gel electrophoresis shows a stabilized native state for tsf:F170L coat proteins. The F170L su substitution also increases procapsid production compared with their tsf parents. We propose that the F170L su substitution has a decreased requirement for the chaperones GroEL and GroES as a result of restoring the tsf coat proteins to a WT-like state. Our data also suggest that GroEL/S can be induced by increasing the population of unfolding intermediates.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
